Histone deacetylase activity regulates chemical diversity in Aspergillus
- PMID: 17616629
- PMCID: PMC2043372
- DOI: 10.1128/EC.00186-07
Histone deacetylase activity regulates chemical diversity in Aspergillus
Abstract
Bioactive small molecules are critical in Aspergillus species during their development and interaction with other organisms. Genes dedicated to their production are encoded in clusters that can be located throughout the genome. We show that deletion of hdaA, encoding an Aspergillus nidulans histone deacetylase (HDAC), causes transcriptional activation of two telomere-proximal gene clusters--and subsequent increased levels of the corresponding molecules (toxin and antibiotic)--but not of a telomere-distal cluster. Introduction of two additional HDAC mutant alleles in a DeltahdaA background had minimal effects on expression of the two HdaA-regulated clusters. Treatment of other fungal genera with HDAC inhibitors resulted in overproduction of several metabolites, suggesting a conserved mechanism of HDAC repression of some secondary-metabolite gene clusters. Chromatin regulation of small-molecule gene clusters may enable filamentous fungi to successfully exploit environmental resources by modifying chemical diversity.
Figures




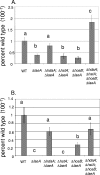
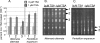
References
-
- Bok, J. W., D. Hoffmeister, L. A. Maggio-Hall, R. Murillo, J. D. Glasner, and N. P. Keller. 2006. Genomic mining for Aspergillus natural products. Chem. Biol. 13:31-37. - PubMed
-
- Bok, J. W., D. Noordermeer, S. P. Kale, and N. P. Keller. 2006. Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol. Microbiol. 61:1636-1645. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

