The TORMOZ gene encodes a nucleolar protein required for regulated division planes and embryo development in Arabidopsis
- PMID: 17616738
- PMCID: PMC1955705
- DOI: 10.1105/tpc.106.042697
The TORMOZ gene encodes a nucleolar protein required for regulated division planes and embryo development in Arabidopsis
Abstract
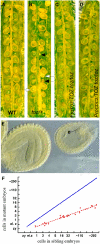
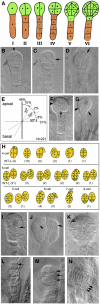
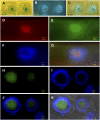
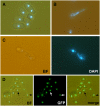
Embryogenesis in Arabidopsis thaliana is marked by a predictable sequence of oriented cell divisions, which precede cell fate determination. We show that mutation of the TORMOZ (TOZ) gene yields embryos with aberrant cell division planes and arrested embryos that appear not to have established normal patterning. The defects in toz mutants differ from previously described mutations that affect embryonic cell division patterns. Longitudinal division planes of the proembryo are frequently replaced by transverse divisions and less frequently by oblique divisions, while divisions of the suspensor cells, which divide only transversely, appear generally unaffected. Expression patterns of selected embryo patterning genes are altered in the mutant embryos, implying that the positional cues required for their proper expression are perturbed by the misoriented divisions. The TOZ gene encodes a nucleolar protein containing WD repeats. Putative TOZ orthologs exist in other eukaryotes including Saccharomyces cerevisiae, where the protein is predicted to function in 18S rRNA biogenesis. We find that disruption of the Sp TOZ gene results in cell division defects in Schizosaccharomyces pombe. Previous studies in yeast and animal cells have identified nucleolar proteins that regulate the exit from M phase and cytokinesis, including factors involved in pre-rRNA processing. Our study suggests that in plant cells, nucleolar functions might interact with the processes of regulated cell divisions and influence the selection of longitudinal division planes during embryogenesis.
Figures




References
-
- Andersen, J.S., Lyon, C.E., Fox, A.H., Leung, A.K., Lam, Y.W., Steen, H., Mann, M., and Lamond, A.I. (2002). Directed proteomic analysis of the human nucleolus. Curr. Biol. 12 1–11. - PubMed
-
- Barneche, F., Steinmetz, F., and Echeverria, M. (2000). Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275 27212–27220. - PubMed
-
- Basi, G., Schmid, E., and Maundrell, K. (1993). TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123 131–136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

