A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes
- PMID: 17616975
- PMCID: PMC1904472
- DOI: 10.1371/journal.ppat.0030095
A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes
Abstract
While the circumstances surrounding the origin and spread of HIV are becoming clearer, the particulars of the origin of simian immunodeficiency virus (SIV) are still unknown. Specifically, the age of SIV, whether it is an ancient or recent infection, has not been resolved. Although many instances of cross-species transmission of SIV have been documented, the similarity between the African green monkey (AGM) and SIVagm phylogenies has long been held as suggestive of ancient codivergence between SIVs and their primate hosts. Here, we present well-resolved phylogenies based on full-length AGM mitochondrial genomes and seven previously published SIVagm genomes; these allowed us to perform the first rigorous phylogenetic test to our knowledge of the hypothesis that SIVagm codiverged with the AGMs. Using the Shimodaira-Hasegawa test, we show that the AGM mitochondrial genomes and SIVagm did not evolve along the same topology. Furthermore, we demonstrate that the SIVagm topology can be explained by a pattern of west-to-east transmission of the virus across existing AGM geographic ranges. Using a relaxed molecular clock, we also provide a date for the most recent common ancestor of the AGMs at approximately 3 million years ago. This study substantially weakens the theory of ancient SIV infection followed by codivergence with its primate hosts.
Conflict of interest statement
Figures
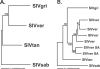

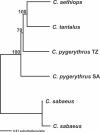
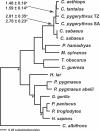
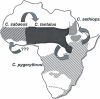
References
-
- McGeoch DJ, Cook S. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J Mol Biol. 1994;238:9–22. - PubMed
-
- Switzer WM, Salemi M, Shanmugam V, Gao F, Cong ME, et al. Ancient co-speciation of simian foamy viruses and primates. Nature. 2005;434:376–380. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

