Evolution of hydra, a recently evolved testis-expressed gene with nine alternative first exons in Drosophila melanogaster
- PMID: 17616977
- PMCID: PMC1904467
- DOI: 10.1371/journal.pgen.0030107
Evolution of hydra, a recently evolved testis-expressed gene with nine alternative first exons in Drosophila melanogaster
Abstract
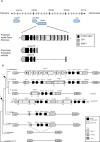
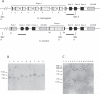
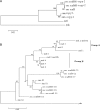
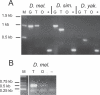
We describe here the Drosophila gene hydra that appears to have originated de novo in the melanogaster subgroup and subsequently evolved in both structure and expression level in Drosophila melanogaster and its sibling species. D. melanogaster hydra encodes a predicted protein of approximately 300 amino acids with no apparent similarity to any previously known proteins. The syntenic region flanking hydra on both sides is found in both D. ananassae and D. pseudoobscura, but hydra is found only in melanogaster subgroup species, suggesting that it originated less than approximately 13 million y ago. Exon 1 of hydra has undergone recurrent duplications, leading to the formation of nine tandem alternative exon 1s in D. melanogaster. Seven of these alternative exons are flanked on their 3' side by the transposon DINE-1 (Drosophila interspersed element-1). We demonstrate that at least four of the nine duplicated exon 1s can function as alternative transcription start sites. The entire hydra locus has also duplicated in D. simulans and D. sechellia. D. melanogaster hydra is expressed most intensely in the proximal testis, suggesting a role in late-stage spermatogenesis. The coding region of hydra has a relatively high Ka/Ks ratio between species, but the ratio is less than 1 in all comparisons, suggesting that hydra is subject to functional constraint. Analysis of sequence polymorphism and divergence of hydra shows that it has evolved under positive selection in the lineage leading to D. melanogaster. The dramatic structural changes surrounding the first exons do not affect the tissue specificity of gene expression: hydra is expressed predominantly in the testes in D. melanogaster, D. simulans, and D. yakuba. However, we have found that expression level changed dramatically (approximately >20-fold) between D. melanogaster and D. simulans. While hydra initially evolved in the absence of nearby transposable element insertions, we suggest that the subsequent accumulation of repetitive sequences in the hydra region may have contributed to structural and expression-level evolution by inducing rearrangements and causing local heterochromatinization. Our analysis further shows that recurrent evolution of both gene structure and expression level may be characteristics of newly evolved genes. We also suggest that late-stage spermatogenesis is the functional target for newly evolved and rapidly evolving male-specific genes.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


References
-
- Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. - PubMed
-
- Long M, Betran E, Thornton K, Wang W. The origin of new genes: Glimpses from the young and old. Nat Rev Gen. 2003;4:865–875. - PubMed
-
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. - PubMed
-
- Doolittle RF. The multiplicity of domains in proteins. Ann Rev Biochem. 1995;64:287–314. - PubMed
-
- Hayashi Y, Sakata H, Makino Y, Urabe I, Yomo T. Can an arbitrary sequence evolve towards acquiring a biological function? J Mol Evol. 2003;56:162–168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

