Sensitivity analysis of intracellular signaling pathway kinetics predicts targets for stem cell fate control
- PMID: 17616983
- PMCID: PMC1913098
- DOI: 10.1371/journal.pcbi.0030130
Sensitivity analysis of intracellular signaling pathway kinetics predicts targets for stem cell fate control
Abstract
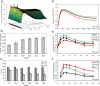
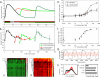
Directing stem cell fate requires knowledge of how signaling networks integrate temporally and spatially segregated stimuli. We developed and validated a computational model of signal transducer and activator of transcription-3 (Stat3) pathway kinetics, a signaling network involved in embryonic stem cell (ESC) self-renewal. Our analysis identified novel pathway responses; for example, overexpression of the receptor glycoprotein-130 results in reduced pathway activation and increased ESC differentiation. We used a systematic in silico screen to identify novel targets and protein interactions involved in Stat3 activation. Our analysis demonstrates that signaling activation and desensitization (the inability to respond to ligand restimulation) is regulated by balancing the activation state of a distributed set of parameters including nuclear export of Stat3, nuclear phosphatase activity, inhibition by suppressor of cytokine signaling, and receptor trafficking. This knowledge was used to devise a temporally modulated ligand delivery strategy that maximizes signaling activation and leads to enhanced ESC self-renewal.
Conflict of interest statement
Figures



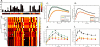
References
-
- Davey RE, Onishi K, Mahdavi A, Zandstra PW. LIF-mediated control of embryonic stem cell self-renewal emerges due to an autoregulatory loop. FASEB J. 2007. Epub March 13. - PubMed
-
- Loebel DA, Watson CM, De Young RA, Tam PP. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev Biol. 2003;264:1–14. - PubMed
-
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. - PubMed
-
- Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

