Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH
- PMID: 17617421
- PMCID: PMC1976599
- DOI: 10.1016/j.jmb.2007.06.034
Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH
Abstract
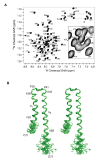
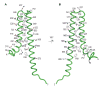
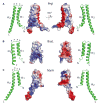
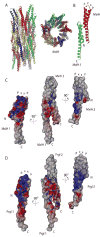
Gram-negative bacteria use a needle-like protein assembly, the type III secretion apparatus, to inject virulence factors into target cells to initiate human disease. The needle is formed by the polymerization of approximately 120 copies of a small acidic protein that is conserved among diverse pathogens. We previously reported the structure of the BsaL needle monomer from Burkholderia pseudomallei by nuclear magnetic resonance (NMR) spectroscopy and others have determined the crystal structure of the Shigella flexneri MxiH needle. Here, we report the NMR structure of the PrgI needle protein of Salmonella typhimurium, a human pathogen associated with food poisoning. PrgI, BsaL, and MxiH form similar two helix bundles, however, the electrostatic surfaces of PrgI differ radically from those of BsaL or MxiH. In BsaL and MxiH, a large negative area is on a face formed by the helix alpha1-alpha2 interface. In PrgI, the major negatively charged surface is not on the "face" but instead is on the "side" of the two-helix bundle, and only residues from helix alpha1 contribute to this negative region. Despite being highly acidic proteins, these molecules contain large basic regions, suggesting that electrostatic contacts are important in needle assembly. Our results also suggest that needle-packing interactions may be different among these bacteria and provide the structural basis for why PrgI and MxiH, despite 63% sequence identity, are not interchangeable in S. typhimurium and S. flexneri.
Figures
Similar articles
-
The common structural architecture of Shigella flexneri and Salmonella typhimurium type three secretion needles.PLoS Pathog. 2013 Mar;9(3):e1003245. doi: 10.1371/journal.ppat.1003245. Epub 2013 Mar 21. PLoS Pathog. 2013. PMID: 23555258 Free PMC article.
-
Physical characterization of MxiH and PrgI, the needle component of the type III secretion apparatus from Shigella and Salmonella.Protein Sci. 2006 Mar;15(3):543-52. doi: 10.1110/ps.051733506. Protein Sci. 2006. PMID: 16501225 Free PMC article.
-
NMR model of PrgI-SipD interaction and its implications in the needle-tip assembly of the Salmonella type III secretion system.J Mol Biol. 2014 Aug 12;426(16):2958-69. doi: 10.1016/j.jmb.2014.06.009. Epub 2014 Jun 18. J Mol Biol. 2014. PMID: 24951833 Free PMC article.
-
Structures of Type III Secretion System Needle Filaments.Curr Top Microbiol Immunol. 2020;427:109-131. doi: 10.1007/82_2019_192. Curr Top Microbiol Immunol. 2020. PMID: 31974760 Review.
-
Structure and biophysics of type III secretion in bacteria.Biochemistry. 2013 Apr 16;52(15):2508-17. doi: 10.1021/bi400160a. Epub 2013 Apr 5. Biochemistry. 2013. PMID: 23521714 Free PMC article. Review.
Cited by
-
Human NAIP/NLRC4 and NLRP3 inflammasomes detect Salmonella type III secretion system activities to restrict intracellular bacterial replication.PLoS Pathog. 2022 Jan 24;18(1):e1009718. doi: 10.1371/journal.ppat.1009718. eCollection 2022 Jan. PLoS Pathog. 2022. PMID: 35073381 Free PMC article.
-
The inner rod protein controls substrate switching and needle length in a Salmonella type III secretion system.Proc Natl Acad Sci U S A. 2014 Jan 14;111(2):817-22. doi: 10.1073/pnas.1319698111. Epub 2013 Dec 30. Proc Natl Acad Sci U S A. 2014. PMID: 24379359 Free PMC article.
-
Structure of AscE and induced burial regions in AscE and AscG upon formation of the chaperone needle-subunit complex of type III secretion system in Aeromonas hydrophila.Protein Sci. 2008 Oct;17(10):1748-60. doi: 10.1110/ps.036798.108. Epub 2008 Jul 28. Protein Sci. 2008. PMID: 18662905 Free PMC article.
-
Helical reconstruction of Salmonella and Shigella needle filaments attached to type 3 basal bodies.Biochem Biophys Rep. 2021 Jun 27;27:101039. doi: 10.1016/j.bbrep.2021.101039. eCollection 2021 Sep. Biochem Biophys Rep. 2021. PMID: 34258394 Free PMC article.
-
Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria.Microbiol Mol Biol Rev. 2012 Jun;76(2):262-310. doi: 10.1128/MMBR.05017-11. Microbiol Mol Biol Rev. 2012. PMID: 22688814 Free PMC article. Review.
References
-
- Vugia DJ, Samuel M, Farley MM, Marcus R, Shiferaw B, Shallow S, Smith K, Angulo FJ. Invasive Salmonella infections in the United States, FoodNet, 1996-1999: incidence, serotype distribution, and outcome. Clin Infect Dis. 2004;38(Suppl 3):S149–S156. - PubMed
-
- Stevens MP, Wood MW, Taylor LA, Monaghan P, Hawes P, Jones PW, Wallis TS, Galyov EE. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol Microbiol. 2002;46:649–659. - PubMed
-
- Yahr TL, Goranson J, Frank DW. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

