Cysteine-cysteine contact preference leads to target-focusing in protein folding
- PMID: 17617551
- PMCID: PMC1913149
- DOI: 10.1529/biophysj.106.097824
Cysteine-cysteine contact preference leads to target-focusing in protein folding
Abstract
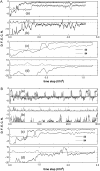
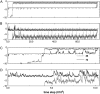
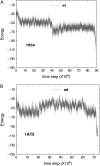
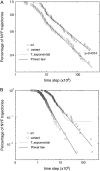
We perform a statistical analysis of amino-acid contacts to investigate possible preferences of amino-acid interactions. We include in the analysis only tertiary contacts, because they are less constrained--compared to secondary contacts--by proteins' backbone rigidity. Using proteins from the protein data bank, our analysis reveals an unusually high frequency of cysteine pairings relative to that expected from random. To elucidate the possible effects of cysteine interactions in folding, we perform molecular simulations on three cysteine-rich proteins. In particular, we investigate the difference in folding dynamics between a Gō-like model (where attraction only occurs between amino acids forming a native contact) and a variant model (where attraction between any two cysteines is introduced to mimic the formation/dissociation of native/nonnative disulfide bonds). We find that when attraction among cysteines is nonspecific and comparable to a solvent-averaged interaction, they produce a target-focusing effect that expedites folding of cysteine-rich proteins as a result of a reduction of conformational search space. In addition, the target-focusing effect also helps reduce glassiness by lowering activation energy barriers and kinetic frustration in the system. The concept of target-focusing also provides a qualitative understanding of a correlation between the rates of protein folding and parameters such as contact order and total contact distance.
Figures
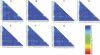
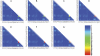
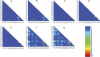
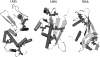


Similar articles
-
Contact pair dynamics during folding of two small proteins: chicken villin head piece and the Alzheimer protein beta-amyloid.J Chem Phys. 2004 Jan 15;120(3):1602-12. doi: 10.1063/1.1633253. J Chem Phys. 2004. PMID: 15268287
-
One short cysteine-rich sequence pattern - two different disulfide-bonded structures - a molecular dynamics simulation study.J Pept Sci. 2015 Jun;21(6):480-94. doi: 10.1002/psc.2765. Epub 2015 Mar 17. J Pept Sci. 2015. PMID: 25781269
-
Importance of native-state topology for determining the folding rate of two-state proteins.J Chem Inf Comput Sci. 2003 Sep-Oct;43(5):1481-5. doi: 10.1021/ci0340308. J Chem Inf Comput Sci. 2003. PMID: 14502481
-
Inter-residue interactions in protein folding and stability.Prog Biophys Mol Biol. 2004 Oct;86(2):235-77. doi: 10.1016/j.pbiomolbio.2003.09.003. Prog Biophys Mol Biol. 2004. PMID: 15288760 Review.
-
Frustration, function and folding.Curr Opin Struct Biol. 2018 Feb;48:68-73. doi: 10.1016/j.sbi.2017.09.006. Epub 2017 Nov 5. Curr Opin Struct Biol. 2018. PMID: 29101782 Free PMC article. Review.
Cited by
-
Computational analysis of C-reactive protein for assessment of molecular dynamics and interaction properties.Cell Biochem Biophys. 2013 Nov;67(2):645-56. doi: 10.1007/s12013-013-9553-4. Cell Biochem Biophys. 2013. PMID: 23494263 Free PMC article.
-
Amino acid patterns around disulfide bonds.Int J Mol Sci. 2010 Nov 18;11(11):4673-86. doi: 10.3390/ijms11114673. Int J Mol Sci. 2010. PMID: 21151463 Free PMC article.
-
A theoretical study of the unfolding pathway of reduced human serum albumin.J Mol Model. 2015 May;21(5):106. doi: 10.1007/s00894-015-2659-4. Epub 2015 Apr 8. J Mol Model. 2015. PMID: 25851108
-
Possible diversifying selection in the imprinted gene, MEDEA, in Arabidopsis.Mol Biol Evol. 2009 Apr;26(4):843-57. doi: 10.1093/molbev/msp001. Epub 2009 Jan 6. Mol Biol Evol. 2009. PMID: 19126870 Free PMC article.
References
-
- Fersht, A. R. 1999. Structure and Mechanism in Protein Science. W. H. Freeman and Company, New York.
-
- Creighton, T. E. 1992. Protein Folding. W. H. Freeman and Company, New York.
-
- Miyazawa, S., and R. L. Jernigan. 1996. Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J. Mol. Biol. 256:623–644. - PubMed
-
- Abkevich, V. I., and E. I. Shakhnovich. 2000. What can disulfide bonds tell us about protein energetics, function and folding: simulations and bioinformatics analysis. J. Mol. Biol. 300:975–985. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

