Analytical biochemistry of DNA--protein assemblies from crude cell extracts
- PMID: 17617645
- PMCID: PMC1935021
- DOI: 10.1093/nar/gkm490
Analytical biochemistry of DNA--protein assemblies from crude cell extracts
Abstract
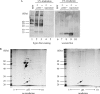
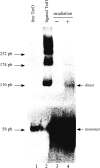
Purification of specific DNA-protein complexes is a challenging task, as the involved interactions can be both electrostatic/H-bond and hydrophobic. The chromatographic stringency needed to obtain reasonable purifications uses salts and detergents. However, these components elicit the removal of proteins unspecifically bound to the chromatographic support itself, thus contaminating the purification products. In this work, a photocleavable linker connected the target oligonucleotidic sequence to the chromatographic beads so as to allow the irradiation-based release of the purified DNA-protein complexes off the beads. Our bioanalytical conditions were validated by purifying the tetracycline repressor protein onto a specific oligonucleotide. The purification factor was unprecedented, with a single contaminant. The robustness of our method was challenged by applying it to the purification of multiprotein assemblies forming onto DNA damage-mimicking oligonucleotides. The purified components were identified as well-known DNA repair proteins, and were shown to retain their enzymatic activities, as seen by monitoring DNA ligation products. Remarkably, kinase activities, also monitored, were found to be distinct on the beads and on the purified DNA-protein complexes, showing the benefits to uncouple the DNA-protein assemblies from the beads for a proper understanding of biochemical regulatory mechanisms involved in the DNA-protein assemblies.
Figures




Similar articles
-
Purification of sequence-specific DNA-binding proteins by affinity chromatography.Curr Protoc Protein Sci. 2001 May;Chapter 9:Unit 9.6. doi: 10.1002/0471140864.ps0906s11. Curr Protoc Protein Sci. 2001. PMID: 18429215
-
Fast gel electrophoresis to analyze DNA-protein interactions.Biotechniques. 1990 May;8(5):556-63. Biotechniques. 1990. PMID: 2357378
-
Tethered bandshift assay and affinity purification of a new DNA-binding protein.Biotechniques. 1994 May;16(5):852-5. Biotechniques. 1994. PMID: 8068340
-
Interaction proteomics: characterization of protein complexes using tandem affinity purification-mass spectrometry.Biochem Soc Trans. 2010 Aug;38(4):883-7. doi: 10.1042/BST0380883. Biochem Soc Trans. 2010. PMID: 20658971 Review.
-
Purification of sequence-specific DNA-binding proteins by affinity chromatography.Curr Protoc Mol Biol. 2001 May;Chapter 12:Unit 12.10. doi: 10.1002/0471142727.mb1210s24. Curr Protoc Mol Biol. 2001. PMID: 18265082
Cited by
-
Unbiased proteomic analysis of proteins interacting with the HIV-1 5'LTR sequence: role of the transcription factor Meis.Nucleic Acids Res. 2012 Nov;40(21):e168. doi: 10.1093/nar/gks733. Epub 2012 Aug 16. Nucleic Acids Res. 2012. PMID: 22904091 Free PMC article.
-
The human DNA ends proteome uncovers an unexpected entanglement of functional pathways.Nucleic Acids Res. 2016 Jun 2;44(10):4721-33. doi: 10.1093/nar/gkw121. Epub 2016 Feb 25. Nucleic Acids Res. 2016. PMID: 26921407 Free PMC article.
-
Protein-DNA/RNA Interactions: An Overview of Investigation Methods in the -Omics Era.J Proteome Res. 2021 Jun 4;20(6):3018-3030. doi: 10.1021/acs.jproteome.1c00074. Epub 2021 May 7. J Proteome Res. 2021. PMID: 33961438 Free PMC article. Review.
References
-
- Rusconi F, Guillonneau F, Praseuth D. Contributions of mass spectrometry in the study of nucleic acid-binding proteins and of nucleic acid-protein interactions. Mass Spectrom. Rev. 2002;21:305–348. - PubMed
-
- Yaneva M, Tempst P. Affinity capture of specific DNA-binding proteins for mass spectrometric identification. Anal. Chem. 2003;75:6437–6448. - PubMed
-
- Yaneva M, Kippenberger S, Wang N, Su Q, McGarvey M, Nazarian A, Lacomis L, Erdjument-Bromage H, Tempst P. PU.1 and a TTTAAA element in the myeloid defensin-1 promoter create an operational TATA Box that can impose cell specificity onto TFIID function. J. Immunol. 2006;176:6906–6917. - PubMed
-
- Nordhoff E, Krogsdam AM, Jorgensen HF, Kallipolitis BH, Clark BF, Roepstorff P, Kristiansen K. Rapid identification of DNA-binding proteins by mass spectrometry. Nat. Biotechnol. 1999;17:884–888. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

