Modeling Neisseria meningitidis metabolism: from genome to metabolic fluxes
- PMID: 17617894
- PMCID: PMC2323225
- DOI: 10.1186/gb-2007-8-7-r136
Modeling Neisseria meningitidis metabolism: from genome to metabolic fluxes
Abstract
Background: Neisseria meningitidis is a human pathogen that can infect diverse sites within the human host. The major diseases caused by N. meningitidis are responsible for death and disability, especially in young infants. In general, most of the recent work on N. meningitidis focuses on potential antigens and their functions, immunogenicity, and pathogenicity mechanisms. Very little work has been carried out on Neisseria primary metabolism over the past 25 years.
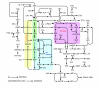
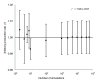
Results: Using the genomic database of N. meningitidis serogroup B together with biochemical and physiological information in the literature we constructed a genome-scale flux model for the primary metabolism of N. meningitidis. The validity of a simplified metabolic network derived from the genome-scale metabolic network was checked using flux-balance analysis in chemostat cultures. Several useful predictions were obtained from in silico experiments, including substrate preference. A minimal medium for growth of N. meningitidis was designed and tested successfully in batch and chemostat cultures.
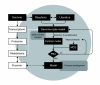
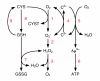
Conclusion: The verified metabolic model describes the primary metabolism of N. meningitidis in a chemostat in steady state. The genome-scale model is valuable because it offers a framework to study N. meningitidis metabolism as a whole, or certain aspects of it, and it can also be used for the purpose of vaccine process development (for example, the design of growth media). The flux distribution of the main metabolic pathways (that is, the pentose phosphate pathway and the Entner-Douderoff pathway) indicates that the major part of pyruvate (69%) is synthesized through the ED-cleavage, a finding that is in good agreement with literature.
Figures
References
-
- Finne J, Bitter-Suermann D, Goridis C, Finne U. An IgG monoclonal antibody to group B meningococci cross-reacts with developmentally regulated polysialic acid units of glycoproteins in neural and extraneural tissues. J Immunol. 1987;138:4402–4407. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

