Analysis of the immune-related transcriptome of a lophotrochozoan model, the marine annelid Platynereis dumerilii
- PMID: 17617895
- PMCID: PMC1939704
- DOI: 10.1186/1742-9994-4-18
Analysis of the immune-related transcriptome of a lophotrochozoan model, the marine annelid Platynereis dumerilii
Abstract
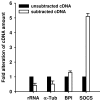
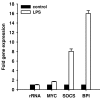
Background: The marine annelid Platynereis dumerilii (Polychaeta, Nereididae) has been recognized as a slow-evolving lophotrochozoan that attracts increasing attention as a valuable model for evolutionary and developmental research. Here, we analyzed its immune-related transcriptome. For targeted identification of immune-induced genes we injected bacterial lipopolysaccharide, a commonly used elicitor of innate immune responses, and applied the suppression subtractive hybridization technique that selectively amplifies cDNAs of differentially expressed genes.
Results: Sequence analysis of 288 cDNAs revealed induced expression of numerous genes whose potential homologues from other animals mediate recognition of infection (e.g. complement receptor CD35), signaling (e.g. myc and SOCS), or act as effector molecules like ferritins and the bactericidal permeability-increasing protein. Interestingly, phylogenetic analyses implicate that immune-related genes identified in P. dumerilii are more related to counterparts from Deuterostomia than are those from Ecdysozoa, similarly as recently described for opsin and intron-rich genes.
Conclusion: Obtained results may allow for a better understanding of Platynereis immunity and support the view that P. dumerilii represents a suitable model for analyzing immune responses of Lophotrochozoa.
Figures





Similar articles
-
PdumBase: a transcriptome database and research tool for Platynereis dumerilii and early development of other metazoans.BMC Genomics. 2018 Aug 16;19(1):618. doi: 10.1186/s12864-018-4987-0. BMC Genomics. 2018. PMID: 30115014 Free PMC article.
-
The neuropeptide complement of the marine annelid Platynereis dumerilii.BMC Genomics. 2013 Dec 20;14:906. doi: 10.1186/1471-2164-14-906. BMC Genomics. 2013. PMID: 24359412 Free PMC article.
-
Structure, phylogeny, and expression of the frizzled-related gene family in the lophotrochozoan annelid Platynereis dumerilii.Evodevo. 2015 Dec 4;6:37. doi: 10.1186/s13227-015-0032-4. eCollection 2015. Evodevo. 2015. PMID: 26640641 Free PMC article.
-
Linking micro- and macro-evolution at the cell type level: a view from the lophotrochozoan Platynereis dumerilii.Brief Funct Genomics. 2013 Sep;12(5):430-9. doi: 10.1093/bfgp/els049. Epub 2012 Nov 20. Brief Funct Genomics. 2013. PMID: 23172798 Review.
-
Genetic and genomic tools for the marine annelid Platynereis dumerilii.Genetics. 2014 May;197(1):19-31. doi: 10.1534/genetics.112.148254. Genetics. 2014. PMID: 24807110 Free PMC article. Review.
Cited by
-
Host Defense Proteins and Peptides with Lipopolysaccharide-Binding Activity from Marine Invertebrates and Their Therapeutic Potential in Gram-Negative Sepsis.Mar Drugs. 2023 Nov 7;21(11):581. doi: 10.3390/md21110581. Mar Drugs. 2023. PMID: 37999405 Free PMC article. Review.
-
Predicted class-I aminoacyl tRNA synthetase-like proteins in non-ribosomal peptide synthesis.Biol Direct. 2010 Aug 2;5:48. doi: 10.1186/1745-6150-5-48. Biol Direct. 2010. PMID: 20678224 Free PMC article.
-
A transcriptional blueprint for a spiral-cleaving embryo.BMC Genomics. 2016 Aug 5;17:552. doi: 10.1186/s12864-016-2860-6. BMC Genomics. 2016. PMID: 27496340 Free PMC article.
-
A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella.BMC Genomics. 2011 Jun 11;12:308. doi: 10.1186/1471-2164-12-308. BMC Genomics. 2011. PMID: 21663692 Free PMC article.
-
Parental transfer of the antimicrobial protein LBP/BPI protects Biomphalaria glabrata eggs against oomycete infections.PLoS Pathog. 2013;9(12):e1003792. doi: 10.1371/journal.ppat.1003792. Epub 2013 Dec 19. PLoS Pathog. 2013. PMID: 24367257 Free PMC article.
References
-
- Cooper EL. Comparative immunology. Integrative Zoology. 2006;1:32–43. - PubMed
LinkOut - more resources
Full Text Sources

