The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities
- PMID: 17617913
- PMCID: PMC1948888
- DOI: 10.1186/1743-422X-4-69
The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities
Abstract
Background: RNA viruses have been isolated that infect marine organisms ranging from bacteria to whales, but little is known about the composition and population structure of the in situ marine RNA virus community. In a recent study, the majority of three genomes of previously unknown positive-sense single-stranded (ss) RNA viruses were assembled from reverse-transcribed whole-genome shotgun libraries. The present contribution comparatively analyzes these genomes with respect to representative viruses from established viral taxa.
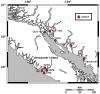
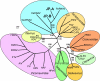
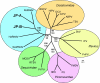
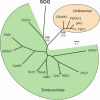
Results: Two of the genomes (JP-A and JP-B), appear to be polycistronic viruses in the proposed order Picornavirales that fall into a well-supported clade of marine picorna-like viruses, the characterized members of which all infect marine protists. A temporal and geographic survey indicates that the JP genomes are persistent and widespread in British Columbia waters. The third genome, SOG, encodes a putative RNA-dependent RNA polymerase (RdRp) that is related to the RdRp of viruses in the family Tombusviridae, but the remaining SOG sequence has no significant similarity to any sequences in the NCBI database.
Conclusion: The complete genomes of these viruses permitted analyses that resulted in a more comprehensive comparison of these pathogens with established taxa. For example, in concordance with phylogenies based on the RdRp, our results support a close homology between JP-A and JP-B and RsRNAV. In contrast, although classification of the SOG genome based on the RdRp places SOG within the Tombusviridae, SOG lacks a capsid and movement protein conserved within this family and SOG is thus likely more distantly related to the Tombusivridae than the RdRp phylogeney indicates.
Figures

Similar articles
-
Metagenomic analysis of coastal RNA virus communities.Science. 2006 Jun 23;312(5781):1795-8. doi: 10.1126/science.1127404. Science. 2006. PMID: 16794078
-
Marine RNA Virus Quasispecies Are Distributed throughout the Oceans.mSphere. 2019 Apr 3;4(2):e00157-19. doi: 10.1128/mSphereDirect.00157-19. mSphere. 2019. PMID: 30944212 Free PMC article.
-
A novel single-stranded RNA virus isolated from the rice-pathogenic fungus Magnaporthe oryzae with similarity to members of the family Tombusviridae.Arch Virol. 2016 Mar;161(3):725-9. doi: 10.1007/s00705-015-2683-9. Epub 2015 Dec 9. Arch Virol. 2016. PMID: 26650038
-
Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences.Crit Rev Biochem Mol Biol. 1993;28(5):375-430. doi: 10.3109/10409239309078440. Crit Rev Biochem Mol Biol. 1993. PMID: 8269709 Review.
-
RNA viruses in the sea.FEMS Microbiol Rev. 2009 Mar;33(2):295-323. doi: 10.1111/j.1574-6976.2008.00132.x. FEMS Microbiol Rev. 2009. PMID: 19243445 Review.
Cited by
-
Virus-derived sequences from the transcriptomes of two snail vectors of schistosomiasis, Biomphalaria pfeifferi and Bulinus globosus from Kenya.PeerJ. 2021 Nov 15;9:e12290. doi: 10.7717/peerj.12290. eCollection 2021. PeerJ. 2021. PMID: 34820163 Free PMC article.
-
Diverse plant viruses: a toolbox for dissection of cellular pathways.J Exp Bot. 2019 Jun 28;70(12):3029-3034. doi: 10.1093/jxb/erz122. J Exp Bot. 2019. PMID: 30882863 Free PMC article.
-
A comprehensive method for amplicon-based and metagenomic characterization of viruses, bacteria, and eukaryotes in freshwater samples.Microbiome. 2016 Jul 19;4(1):20. doi: 10.1186/s40168-016-0166-1. Microbiome. 2016. PMID: 27391119 Free PMC article.
-
Metagenomic analysis of RNA viruses in a fresh water lake.PLoS One. 2009 Sep 29;4(9):e7264. doi: 10.1371/journal.pone.0007264. PLoS One. 2009. PMID: 19787045 Free PMC article.
-
Putative Invertebrate, Plant, and Wastewater Derived ssRNA Viruses in Plankton of the Anthropogenically Impacted Anacostia River, District of Columbia, USA.Microbes Environ. 2022;37(5):ME21070. doi: 10.1264/jsme2.ME21070. Microbes Environ. 2022. PMID: 35264468 Free PMC article.
References
-
- Smith A. Aquatic virus cycles. In: Hurst C, editor. Viral Ecology. San Diego: Academic Press; 2000. pp. 447–491.
-
- Nagasaki K, Tomaru Y, Katanozaka N, Shirai Y, Nishida K, Itakura S, Yamaguchi M. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl Environ Microbiol. 2004;70:704–711. doi: 10.1128/AEM.70.2.704-711.2004. - DOI - PMC - PubMed
-
- Tomaru Y, Katanozaka N, Nishida K, Shirai Y, Tarutani K, Yamaguchi M, Nagasaki K. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat Microb Ecol. 2004;34:207–218.
-
- Tai V, Lawrence JE, Lang AS, Chan AM, Culley AI, Suttle CA. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Hetersigma akashiwo (Raphidophyceae) J Phycol. 2003;39:343–352.
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

