Collective properties of evolving molecular quasispecies
- PMID: 17620110
- PMCID: PMC1934359
- DOI: 10.1186/1471-2148-7-110
Collective properties of evolving molecular quasispecies
Abstract
Background: RNA molecules, through their dual appearance as sequence and structure, represent a suitable model to study evolutionary properties of quasispecies. The essential ingredient in this model is the differentiation between genotype (molecular sequences which are affected by mutation) and phenotype (molecular structure, affected by selection). This framework allows a quantitative analysis of organizational properties of quasispecies as they adapt to different environments, such as their robustness, the effect of the degeneration of the sequence space, or the adaptation under different mutation rates and the error threshold associated.
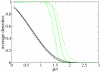
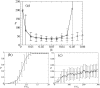
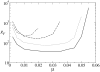
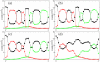
Results: We describe and analyze the structural properties of molecular quasispecies adapting to different environments both during the transient time before adaptation takes place and in the asymptotic state, once optimization has occurred. We observe a minimum in the adaptation time at values of the mutation rate relatively far from the phenotypic error threshold. Through the definition of a consensus structure, it is shown that the quasispecies retains relevant structural information in a distributed fashion even above the error threshold. This structural robustness depends on the precise shape of the secondary structure used as target of selection. Experimental results available for natural RNA populations are in qualitative agreement with our observations.
Conclusion: Adaptation time of molecular quasispecies to a given environment is optimized at values of the mutation rate well below the phenotypic error threshold. The optimal value results from a trade-off between diversity generation and fixation of advantageous mutants. The critical value of the mutation rate is a function not only of the sequence length, but also of the specific properties of the environment, in this case the selection pressure and the shape of the secondary structure used as target phenotype. Certain functional motifs of RNA secondary structure that withstand high mutation rates (as the ubiquitous hairpin motif) might appear early in evolution and be actually frozen evolutionary accidents.
Figures



Similar articles
-
Phenotypic effect of mutations in evolving populations of RNA molecules.BMC Evol Biol. 2010 Feb 17;10:46. doi: 10.1186/1471-2148-10-46. BMC Evol Biol. 2010. PMID: 20163698 Free PMC article.
-
RNA based evolutionary optimization.Orig Life Evol Biosph. 1993 Dec;23(5-6):373-91. doi: 10.1007/BF01582087. Orig Life Evol Biosph. 1993. PMID: 7509478
-
Replication and mutation on neutral networks.Bull Math Biol. 2001 Jan;63(1):57-94. doi: 10.1006/bulm.2000.0206. Bull Math Biol. 2001. PMID: 11146884
-
Quasispecies on Fitness Landscapes.Curr Top Microbiol Immunol. 2016;392:61-120. doi: 10.1007/82_2015_469. Curr Top Microbiol Immunol. 2016. PMID: 26597856 Review.
-
Quasispecies made simple.PLoS Comput Biol. 2005 Nov;1(6):e61. doi: 10.1371/journal.pcbi.0010061. PLoS Comput Biol. 2005. PMID: 16322763 Free PMC article. Review.
Cited by
-
On the emergence of structural complexity in RNA replicators.RNA. 2019 Dec;25(12):1579-1591. doi: 10.1261/rna.070391.119. Epub 2019 Aug 29. RNA. 2019. PMID: 31467146 Free PMC article.
-
The advantage of arriving first: characteristic times in finite size populations of error-prone replicators.PLoS One. 2013 Dec 23;8(12):e83142. doi: 10.1371/journal.pone.0083142. eCollection 2013. PLoS One. 2013. PMID: 24376656 Free PMC article.
-
Eco-evolutionary dynamics, coding structure and the information threshold.BMC Evol Biol. 2010 Nov 24;10:361. doi: 10.1186/1471-2148-10-361. BMC Evol Biol. 2010. PMID: 21106077 Free PMC article.
-
Potential benefits of sequential inhibitor-mutagen treatments of RNA virus infections.PLoS Pathog. 2009 Nov;5(11):e1000658. doi: 10.1371/journal.ppat.1000658. Epub 2009 Nov 13. PLoS Pathog. 2009. PMID: 19911056 Free PMC article.
-
Pathways to extinction: beyond the error threshold.Philos Trans R Soc Lond B Biol Sci. 2010 Jun 27;365(1548):1943-52. doi: 10.1098/rstb.2010.0076. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20478889 Free PMC article.
References
-
- Eigen M, Schuster P. The hypercycle A principle of natural self-organization. Springer-Verlag, Heidelberg; 1979. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

