Gene duplication, modularity and adaptation in the evolution of the aflatoxin gene cluster
- PMID: 17620135
- PMCID: PMC1949824
- DOI: 10.1186/1471-2148-7-111
Gene duplication, modularity and adaptation in the evolution of the aflatoxin gene cluster
Abstract
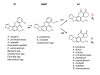
Background: The biosynthesis of aflatoxin (AF) involves over 20 enzymatic reactions in a complex polyketide pathway that converts acetate and malonate to the intermediates sterigmatocystin (ST) and O-methylsterigmatocystin (OMST), the respective penultimate and ultimate precursors of AF. Although these precursors are chemically and structurally very similar, their accumulation differs at the species level for Aspergilli. Notable examples are A. nidulans that synthesizes only ST, A. flavus that makes predominantly AF, and A. parasiticus that generally produces either AF or OMST. Whether these differences are important in the evolutionary/ecological processes of species adaptation and diversification is unknown. Equally unknown are the specific genomic mechanisms responsible for ordering and clustering of genes in the AF pathway of Aspergillus.
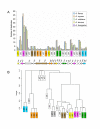
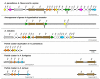
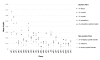
Results: To elucidate the mechanisms that have driven formation of these clusters, we performed systematic searches of aflatoxin cluster homologs across five Aspergillus genomes. We found a high level of gene duplication and identified seven modules consisting of highly correlated gene pairs (aflA/aflB, aflR/aflS, aflX/aflY, aflF/aflE, aflT/aflQ, aflC/aflW, and aflG/aflL). With the exception of A. nomius, contrasts of mean Ka/Ks values across all cluster genes showed significant differences in selective pressure between section Flavi and non-section Flavi species. A. nomius mean Ka/Ks values were more similar to partial clusters in A. fumigatus and A. terreus. Overall, mean Ka/Ks values were significantly higher for section Flavi than for non-section Flavi species.
Conclusion: Our results implicate several genomic mechanisms in the evolution of ST, OMST and AF cluster genes. Gene modules may arise from duplications of a single gene, whereby the function of the pre-duplication gene is retained in the copy (aflF/aflE) or the copies may partition the ancestral function (aflA/aflB). In some gene modules, the duplicated copy may simply augment/supplement a specific pathway function (aflR/aflS and aflX/aflY) or the duplicated copy may evolve a completely new function (aflT/aflQ and aflC/aflW). Gene modules that are contiguous in one species and noncontiguous in others point to possible rearrangements of cluster genes in the evolution of these species. Significantly higher mean Ka/Ks values in section Flavi compared to non-section Flavi species indicate increased positive selection acting in the evolution of genes in OMST and AF gene clusters.
Figures
References
-
- Frisvad JC, Samson RA. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology. 2004;49:1–173.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

