Automatic extraction of gene ontology annotation and its correlation with clusters in protein networks
- PMID: 17620146
- PMCID: PMC1940026
- DOI: 10.1186/1471-2105-8-243
Automatic extraction of gene ontology annotation and its correlation with clusters in protein networks
Abstract
Background: Uncovering cellular roles of a protein is a task of tremendous importance and complexity that requires dedicated experimental work as well as often sophisticated data mining and processing tools. Protein functions, often referred to as its annotations, are believed to manifest themselves through topology of the networks of inter-proteins interactions. In particular, there is a growing body of evidence that proteins performing the same function are more likely to interact with each other than with proteins with other functions. However, since functional annotation and protein network topology are often studied separately, the direct relationship between them has not been comprehensively demonstrated. In addition to having the general biological significance, such demonstration would further validate the data extraction and processing methods used to compose protein annotation and protein-protein interactions datasets.
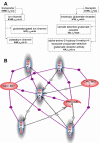
Results: We developed a method for automatic extraction of protein functional annotation from scientific text based on the Natural Language Processing (NLP) technology. For the protein annotation extracted from the entire PubMed, we evaluated the precision and recall rates, and compared the performance of the automatic extraction technology to that of manual curation used in public Gene Ontology (GO) annotation. In the second part of our presentation, we reported a large-scale investigation into the correspondence between communities in the literature-based protein networks and GO annotation groups of functionally related proteins. We found a comprehensive two-way match: proteins within biological annotation groups form significantly denser linked network clusters than expected by chance and, conversely, densely linked network communities exhibit a pronounced non-random overlap with GO groups. We also expanded the publicly available GO biological process annotation using the relations extracted by our NLP technology. An increase in the number and size of GO groups without any noticeable decrease of the link density within the groups indicated that this expansion significantly broadens the public GO annotation without diluting its quality. We revealed that functional GO annotation correlates mostly with clustering in a physical interaction protein network, while its overlap with indirect regulatory network communities is two to three times smaller.
Conclusion: Protein functional annotations extracted by the NLP technology expand and enrich the existing GO annotation system. The GO functional modularity correlates mostly with the clustering in the physical interaction network, suggesting that the essential role of structural organization maintained by these interactions. Reciprocally, clustering of proteins in physical interaction networks can serve as an evidence for their functional similarity.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

