Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles
- PMID: 17620411
- PMCID: PMC2064446
- DOI: 10.1083/jcb.200702141
Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles
Abstract
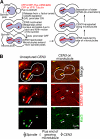
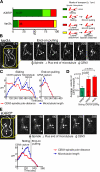
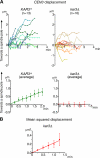
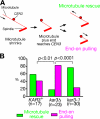
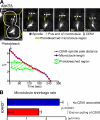
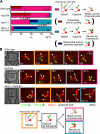
In mitosis, kinetochores are initially captured by the lateral sides of single microtubules and are subsequently transported toward spindle poles. Mechanisms for kinetochore transport are not yet known. We present two mechanisms involved in microtubule-dependent poleward kinetochore transport in Saccharomyces cerevisiae. First, kinetochores slide along the microtubule lateral surface, which is mainly and probably exclusively driven by Kar3, a kinesin-14 family member that localizes at kinetochores. Second, kinetochores are tethered at the microtubule distal ends and pulled poleward as microtubules shrink (end-on pulling). Kinetochore sliding is often converted to end-on pulling, enabling more processive transport, but the opposite conversion is rare. The establishment of end-on pulling is partly hindered by Kar3, and its progression requires the Dam1 complex. We suggest that the Dam1 complexes, which probably encircle a single microtubule, can convert microtubule depolymerization into the poleward kinetochore-pulling force. Thus, microtubule-dependent poleward kinetochore transport is ensured by at least two distinct mechanisms.
Figures




References
-
- Amberg, D.C., D.J. Burke, and J.N. Strathern. 2005. Methods in Yeast Genetics: a Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 230 pp.
-
- Andrews, P.D., Y. Ovechkina, N. Morrice, M. Wagenbach, K. Duncan, L. Wordeman, and J.R. Swedlow. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

