Nucleosome positioning signals in genomic DNA
- PMID: 17620451
- PMCID: PMC1933512
- DOI: 10.1101/gr.6101007
Nucleosome positioning signals in genomic DNA
Abstract
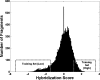
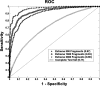
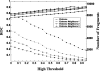
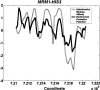
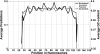
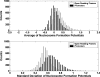
Although histones can form nucleosomes on virtually any genomic sequence, DNA sequences show considerable variability in their binding affinity. We have used DNA sequences of Saccharomyces cerevisiae whose nucleosome binding affinities have been experimentally determined (Yuan et al. 2005) to train a support vector machine to identify the nucleosome formation potential of any given sequence of DNA. The DNA sequences whose nucleosome formation potential are most accurately predicted are those that contain strong nucleosome forming or inhibiting signals and are found within nucleosome length stretches of genomic DNA with continuous nucleosome formation or inhibition signals. We have accurately predicted the experimentally determined nucleosome positions across a well-characterized promoter region of S. cerevisiae and identified strong periodicity within 199 center-aligned mononucleosomes studied recently (Segal et al. 2006) despite there being no periodicity information used to train the support vector machine. Our analysis suggests that only a subset of nucleosomes are likely to be positioned by intrinsic sequence signals. This observation is consistent with the available experimental data and is inconsistent with the proposal of a nucleosome positioning code. Finally, we show that intrinsic nucleosome positioning signals are both more inhibitory and more variable in promoter regions than in open reading frames in S. cerevisiae.
Figures

References
-
- Alberts B. Molecular biology of the cell. Garland Science; New York: 2002.
-
- Anselmi C., Bocchinfuso G., De Santis P., Savino M., Scipioni A., Bocchinfuso G., De Santis P., Savino M., Scipioni A., De Santis P., Savino M., Scipioni A., Savino M., Scipioni A., Scipioni A. Dual role of DNA intrinsic curvature and flexibility in determining nucleosome stability. J. Mol. Biol. 1999;286:1293–1301. - PubMed
-
- Baldi P., Brunak S., Chauvin Y., Krogh A., Brunak S., Chauvin Y., Krogh A., Chauvin Y., Krogh A., Krogh A. Naturally occurring nucleosome positioning signals in human exons and introns. J. Mol. Biol. 1996;263:503–510. - PubMed
-
- Boeger H., Griesenbeck J., Strattan J.S., Kornberg R.D., Griesenbeck J., Strattan J.S., Kornberg R.D., Strattan J.S., Kornberg R.D., Kornberg R.D. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell. 2003;11:1587–1598. - PubMed
-
- Boeger H., Griesenbeck J., Strattan J.S., Kornberg R.D., Griesenbeck J., Strattan J.S., Kornberg R.D., Strattan J.S., Kornberg R.D., Kornberg R.D. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell. 2004;14:667–673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
