Directed evolution can rapidly improve the activity of chimeric assembly-line enzymes
- PMID: 17620609
- PMCID: PMC1924594
- DOI: 10.1073/pnas.0705348104
Directed evolution can rapidly improve the activity of chimeric assembly-line enzymes
Abstract
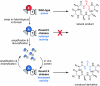
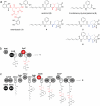
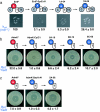
Nonribosomal peptides (NRPs) are produced by NRP synthetase (NRPS) enzymes that function as molecular assembly lines. The modular architecture of NRPSs suggests that a domain responsible for activating a building block could be replaced with a domain from a foreign NRPS to create a chimeric assembly line that produces a new variant of a natural NRP. However, such chimeric NRPS modules are often heavily impaired, impeding efforts to create novel NRP variants by swapping domains from different modules or organisms. Here we show that impaired chimeric NRPSs can be functionally restored by directed evolution. Using rounds of mutagenesis coupled with in vivo screens for NRP production, we rapidly isolated variants of two different chimeric NRPSs with approximately 10-fold improvements in enzyme activity and product yield, including one that produces new derivatives of the potent NRP/polyketide antibiotic andrimid. Because functional restoration in these examples required only modest library sizes (10(3) to 10(4) clones) and three or fewer rounds of screening, our approach may be widely applicable even for NRPSs from genetically challenging hosts.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Sieber SA, Marahiel MA. Chem Rev. 2005;105:715–738. - PubMed
-
- Fischbach MA, Walsh CT. Chem Rev. 2006;106:3468–3496. - PubMed
-
- Baltz RH. Nat Biotechnol. 2006;24:1533–1540. - PubMed
-
- Cane DE, Walsh CT, Khosla C. Science. 1998;282:63–68. - PubMed
-
- Coeffet-Le Gal MF, Thurston L, Rich P, Miao V, Baltz RH. Microbiology. 2006;152:2993–3001. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

