Myc dynamically and preferentially relocates to a transcription factory occupied by Igh
- PMID: 17622196
- PMCID: PMC1945077
- DOI: 10.1371/journal.pbio.0050192
Myc dynamically and preferentially relocates to a transcription factory occupied by Igh
Abstract
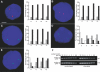
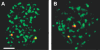
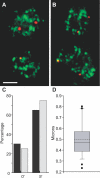
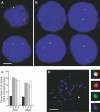
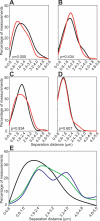
Transcription in mammalian nuclei is highly compartmentalized in RNA polymerase II-enriched nuclear foci known as transcription factories. Genes in cis and trans can share the same factory, suggesting that genes migrate to preassembled transcription sites. We used fluorescent in situ hybridization to investigate the dynamics of gene association with transcription factories during immediate early (IE) gene induction in mouse B lymphocytes. Here, we show that induction involves rapid gene relocation to transcription factories. Importantly, we find that the Myc proto-oncogene on Chromosome 15 is preferentially recruited to the same transcription factory as the highly transcribed Igh gene located on Chromosome 12. Myc and Igh are the most frequent translocation partners in plasmacytoma and Burkitt lymphoma. Our results show that transcriptional activation of IE genes involves rapid relocation to preassembled transcription factories. Furthermore, the data imply a direct link between the nonrandom interchromosomal organization of transcribed genes at transcription factories and the incidence of specific chromosomal translocations.
Conflict of interest statement
Figures


References
-
- Cremer M, von Hase J, Volm T, Brero A, Kreth G, et al. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. - PubMed
-
- Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, et al. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

