Evaluation of gene panel mRNAs in urine samples of kidney transplant recipients as a non-invasive tool of graft function
- PMID: 17622313
- PMCID: PMC1906687
- DOI: 10.2119/2007–00017.Mas
Evaluation of gene panel mRNAs in urine samples of kidney transplant recipients as a non-invasive tool of graft function
Abstract
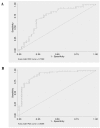
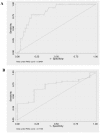
Non-invasive monitoring may be useful after kidney transplantation (KT), particularly for predicting acute rejection (AR). It is less clear whether chronic allograft nephropathy (CAN) is also associated with changes in urine cells. To identify non-invasive markers of allograft function in kidney transplant patients (KTP), mRNA levels of AGT, TGF-beta1, EGFR, IFN-gamma, TSP-1, and IL-10 in urine (Ur) samples were studied using QRT-PCR. Ninety-five KTP and 111 Ur samples were evaluated. Patients (Pts) were divided as, within six months (N = 31), and with more than six months post-KT (N = 64). KTP with more than six months post-KT were classified as KTP with stable kidney function (SKF) (N = 32), KTP with SKF (creatinine < 2 mg/dL) and proteinuria > 500 mg/24 h (N = 18), and KTP with biopsy proven CAN (N = 14). F-test was used to test for equality of variances between groups. IL-10 mRNA was decreased in Ur samples from KTP with less than six months post-KT (P = 0.005). For KTR groups with more than six months post-KT, AGT and EGFR mRNA were statistically different among KTP with SKF, KTP with SKF and proteinuria, and CAN Pts (P = 0.003, and P = 0.01), with KTP with SKF having higher mean expression. TSP-1 mRNA levels also were significantly different among these three groups (P = 0.04), with higher expression observed in CAN Pts. Using the random forest algorithm, AGT, EGFR, and TGF-beta1 were identified as predictors of CAN, SKF, SKF with proteinuria. A characteristic pattern of mRNA levels in the different KTP groups was observed indicating that the mRNA levels in Ur cells might reflect allograft function.
Figures


References
-
- Gjertson DW. Impact of delayed graft function and acute rejection on kidney graft survival. Clin Transpl. 2000;14:467. - PubMed
-
- Cecka JM. The UNOS scientific renal transplant registry. In: Cecka JM, Terasaki PI, editors. Clinical Transplants. UCLA Immunogenetics Center; 1999. 2000. p. 1. - PubMed
-
- Nankivell BJ, Chapman JR. Chronic allograft nephropathy: current concepts and future directions. Transplantation. 2006;81:643–54. - PubMed
-
- Saurina A, et al. Conversion from calcineurin inhibitors to sirolimus in chronic allograft dysfunction: changes in glomerular haemodynamics and proteinuria. Nephrol Dial Transplant. 2006;21:488–93. - PubMed
-
- Kasiske BL, et al. Long-term deterioration of kidney allograft function. Am J Transplant. 2005;5:1405–14. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
