Efficient and specific internal cleavage of a retroviral palindromic DNA sequence by tetrameric HIV-1 integrase
- PMID: 17622353
- PMCID: PMC1905944
- DOI: 10.1371/journal.pone.0000608
Efficient and specific internal cleavage of a retroviral palindromic DNA sequence by tetrameric HIV-1 integrase
Abstract
Background: HIV-1 integrase (IN) catalyses the retroviral integration process, removing two nucleotides from each long terminal repeat and inserting the processed viral DNA into the target DNA. It is widely assumed that the strand transfer step has no sequence specificity. However, recently, it has been reported by several groups that integration sites display a preference for palindromic sequences, suggesting that a symmetry in the target DNA may stabilise the tetrameric organisation of IN in the synaptic complex.
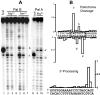
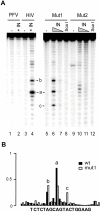
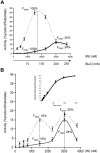
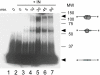
Methodology/principal findings: We assessed the ability of several palindrome-containing sequences to organise tetrameric IN and investigated the ability of IN to catalyse DNA cleavage at internal positions. Only one palindromic sequence was successfully cleaved by IN. Interestingly, this symmetrical sequence corresponded to the 2-LTR junction of retroviral DNA circles-a palindrome similar but not identical to the consensus sequence found at integration sites. This reaction depended strictly on the cognate retroviral sequence of IN and required a full-length wild-type IN. Furthermore, the oligomeric state of IN responsible for this cleavage differed from that involved in the 3'-processing reaction. Palindromic cleavage strictly required the tetrameric form, whereas 3'-processing was efficiently catalysed by a dimer.
Conclusions/significance: Our findings suggest that the restriction-like cleavage of palindromic sequences may be a general physiological activity of retroviral INs and that IN tetramerisation is strongly favoured by DNA symmetry, either at the target site for the concerted integration or when the DNA contains the 2-LTR junction in the case of the palindromic internal cleavage.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

