Structural comparisons of the nucleoprotein from three negative strand RNA virus families
- PMID: 17623082
- PMCID: PMC2031895
- DOI: 10.1186/1743-422X-4-72
Structural comparisons of the nucleoprotein from three negative strand RNA virus families
Abstract
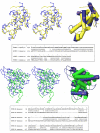
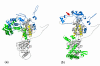
Structures of the nucleoprotein of three negative strand RNA virus families, borna disease virus, rhabdovirus and influenza A virus, are now available. Structural comparisons showed that the topology of the RNA binding region from the three proteins is very similar. The RNA was shown to fit into a cavity formed by the two distinct domains of the RNA binding region in the rhabdovirus nucleoprotein. Two helices connecting the two domains characterize the center of the cavity. The nucleoproteins contain at least 5 conserved helices in the N-terminal domain and 3 conserved helices in the C-terminal domain. Since all negative strand RNA viruses are required to have the ribonucleoprotein complex as their active genomic templates, it is perceivable that the (5H+3H) structure is a common motif in the nucleoprotein of negative strand RNA viruses.
Figures
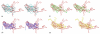
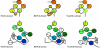
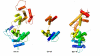

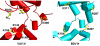
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

