A survey of orphan enzyme activities
- PMID: 17623104
- PMCID: PMC1940265
- DOI: 10.1186/1471-2105-8-244
A survey of orphan enzyme activities
Abstract
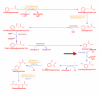
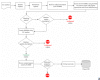
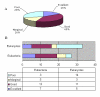
Background: Using computational database searches, we have demonstrated previously that no gene sequences could be found for at least 36% of enzyme activities that have been assigned an Enzyme Commission number. Here we present a follow-up literature-based survey involving a statistically significant sample of such "orphan" activities. The survey was intended to determine whether sequences for these enzyme activities are truly unknown, or whether these sequences are absent from the public sequence databases but can be found in the literature.
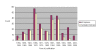
Results: We demonstrate that for ~80% of sampled orphans, the absence of sequence data is bona fide. Our analyses further substantiate the notion that many of these enzyme activities play biologically important roles.
Conclusion: This survey points toward significant scientific cost of having such a large fraction of characterized enzyme activities disconnected from sequence data. It also suggests that a larger effort, beginning with a comprehensive survey of all putative orphan activities, would resolve nearly 300 artifactual orphans and reconnect a wealth of enzyme research with modern genomics. For these reasons, we propose that a systematic effort to identify the cognate genes of orphan enzymes be undertaken.
Figures
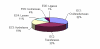
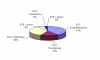
Similar articles
-
Profiling the orphan enzymes.Biol Direct. 2014 Jun 6;9:10. doi: 10.1186/1745-6150-9-10. Biol Direct. 2014. PMID: 24906382 Free PMC article. Review.
-
ORENZA: a web resource for studying ORphan ENZyme activities.BMC Bioinformatics. 2006 Oct 6;7:436. doi: 10.1186/1471-2105-7-436. BMC Bioinformatics. 2006. PMID: 17026747 Free PMC article.
-
Distribution of orphan metabolic activities.Trends Biotechnol. 2007 Aug;25(8):343-8. doi: 10.1016/j.tibtech.2007.06.001. Epub 2007 Jun 18. Trends Biotechnol. 2007. PMID: 17580095
-
The complement of enzymatic sets in different species.J Mol Biol. 2005 Jun 17;349(4):745-63. doi: 10.1016/j.jmb.2005.04.027. J Mol Biol. 2005. PMID: 15896806
-
Orphan enzymes could be an unexplored reservoir of new drug targets.Drug Discov Today. 2006 Apr;11(7-8):300-5. doi: 10.1016/j.drudis.2006.02.002. Drug Discov Today. 2006. PMID: 16580971 Review.
Cited by
-
Rapid identification of sequences for orphan enzymes to power accurate protein annotation.PLoS One. 2013 Dec 30;8(12):e84508. doi: 10.1371/journal.pone.0084508. eCollection 2013. PLoS One. 2013. PMID: 24386392 Free PMC article.
-
Listeria monocytogenes GlmR Is an Accessory Uridyltransferase Essential for Cytosolic Survival and Virulence.mBio. 2023 Apr 25;14(2):e0007323. doi: 10.1128/mbio.00073-23. Epub 2023 Mar 20. mBio. 2023. PMID: 36939339 Free PMC article.
-
Profiling the orphan enzymes.Biol Direct. 2014 Jun 6;9:10. doi: 10.1186/1745-6150-9-10. Biol Direct. 2014. PMID: 24906382 Free PMC article. Review.
-
Finding sequences for over 270 orphan enzymes.PLoS One. 2014 May 14;9(5):e97250. doi: 10.1371/journal.pone.0097250. eCollection 2014. PLoS One. 2014. PMID: 24826896 Free PMC article.
-
The evolution of enzyme function in the isomerases.Curr Opin Struct Biol. 2014 Jun;26:121-30. doi: 10.1016/j.sbi.2014.06.002. Epub 2014 Jul 5. Curr Opin Struct Biol. 2014. PMID: 25000289 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

