Biochemical characterization of a recombinant Japanese encephalitis virus RNA-dependent RNA polymerase
- PMID: 17623110
- PMCID: PMC1934914
- DOI: 10.1186/1471-2199-8-59
Biochemical characterization of a recombinant Japanese encephalitis virus RNA-dependent RNA polymerase
Abstract
Background: Japanese encephalitis virus (JEV) NS5 is a viral nonstructural protein that carries both methyltransferase and RNA-dependent RNA polymerase (RdRp) domains. It is a key component of the viral RNA replicase complex that presumably includes other viral nonstructural and cellular proteins. The biochemical properties of JEV NS5 have not been characterized due to the lack of a robust in vitro RdRp assay system, and the molecular mechanisms for the initiation of RNA synthesis by JEV NS5 remain to be elucidated.
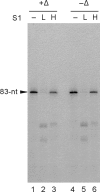
Results: To characterize the biochemical properties of JEV RdRp, we expressed in Escherichia coli and purified an enzymatically active full-length recombinant JEV NS5 protein with a hexahistidine tag at the N-terminus. The purified NS5 protein, but not the mutant NS5 protein with an Ala substitution at the first Asp of the RdRp-conserved GDD motif, exhibited template- and primer-dependent RNA synthesis activity using a poly(A) RNA template. The NS5 protein was able to use both plus- and minus-strand 3'-untranslated regions of the JEV genome as templates in the absence of a primer, with the latter RNA being a better template. Analysis of the RNA synthesis initiation site using the 3'-end 83 nucleotides of the JEV genome as a minimal RNA template revealed that the NS5 protein specifically initiates RNA synthesis from an internal site, U81, at the two nucleotides upstream of the 3'-end of the template.
Conclusion: As a first step toward the understanding of the molecular mechanisms for JEV RNA replication and ultimately for the in vitro reconstitution of viral RNA replicase complex, we for the first time established an in vitro JEV RdRp assay system with a functional full-length recombinant JEV NS5 protein and characterized the mechanisms of RNA synthesis from nonviral and viral RNA templates. The full-length recombinant JEV NS5 will be useful for the elucidation of the structure-function relationship of this enzyme and for the development of anti-JEV agents.
Figures

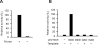
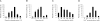


Similar articles
-
Identification and characterization of RNA-dependent RNA polymerase activity in recombinant Japanese encephalitis virus NS5 protein.Arch Virol. 2007;152(10):1859-69. doi: 10.1007/s00705-007-1007-0. Epub 2007 Jun 18. Arch Virol. 2007. PMID: 17577613 Free PMC article.
-
Organization of flaviviral replicase proteins in virus-induced membranes: a role for NS1' in Japanese encephalitis virus RNA synthesis.Novartis Found Symp. 2006;277:136-45; discussion 145-8, 251-3. Novartis Found Symp. 2006. PMID: 17319159
-
Effective inhibition of Japanese encephalitis virus replication by small interfering RNAs targeting the NS5 gene.Virus Res. 2008 Mar;132(1-2):145-51. doi: 10.1016/j.virusres.2007.11.014. Epub 2008 Jan 10. Virus Res. 2008. PMID: 18190994
-
A structural view of the RNA-dependent RNA polymerases from the Flavivirus genus.Virus Res. 2017 Apr 15;234:34-43. doi: 10.1016/j.virusres.2017.01.020. Epub 2017 Jan 25. Virus Res. 2017. PMID: 28131854 Review.
-
RNA-dependent RNA polymerases of dsRNA bacteriophages.Virus Res. 2004 Apr;101(1):45-55. doi: 10.1016/j.virusres.2003.12.005. Virus Res. 2004. PMID: 15010216 Review.
Cited by
-
Influence of Zika virus 3'-end sequence and nonstructural protein evolution on the viral replication competence and virulence.Emerg Microbes Infect. 2022 Dec;11(1):2447-2465. doi: 10.1080/22221751.2022.2128433. Emerg Microbes Infect. 2022. PMID: 36149812 Free PMC article.
-
Galidesivir Triphosphate Promotes Stalling of Dengue-2 Virus Polymerase Immediately Prior to Incorporation.ACS Infect Dis. 2023 Aug 11;9(8):1658-1673. doi: 10.1021/acsinfecdis.3c00311. Epub 2023 Jul 24. ACS Infect Dis. 2023. PMID: 37488090 Free PMC article.
-
Essential role of γ-clade RNA-dependent RNA polymerases in rice development and yield-related traits is linked to their atypical polymerase activities regulating specific genomic regions.New Phytol. 2021 Nov;232(4):1674-1691. doi: 10.1111/nph.17700. Epub 2021 Sep 16. New Phytol. 2021. PMID: 34449900 Free PMC article.
-
Structural and Biochemical Studies on the Reaction Mechanism of Uridine-Cytidine Kinase.Protein J. 2015 Dec;34(6):411-20. doi: 10.1007/s10930-015-9636-8. Protein J. 2015. PMID: 26510656
-
Mechanistic insights into the Japanese encephalitis virus RNA dependent RNA polymerase protein inhibition by bioflavonoids from Azadirachta indica.Sci Rep. 2021 Sep 13;11(1):18125. doi: 10.1038/s41598-021-96917-0. Sci Rep. 2021. PMID: 34518560 Free PMC article.
References
-
- Hanna JN, Ritchie SA, Phillips DA, Shield J, Bailey MC, Mackenzie JS, Poidinger M, McCall BJ, Mills PJ. An outbreak of Japanese encephalitis in the Torres Strait, Australia, 1995. Med J Aust. 1996;165:256–260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

