GNL3L inhibits activity of estrogen-related receptor gamma by competing for coactivator binding
- PMID: 17623774
- PMCID: PMC2975966
- DOI: 10.1242/jcs.009878
GNL3L inhibits activity of estrogen-related receptor gamma by competing for coactivator binding
Abstract
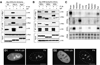
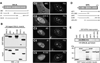
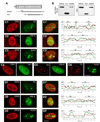
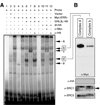
Guanine nucleotide binding protein-like 3 (GNL3L) is the closest homologue of a stem cell-enriched factor nucleostemin in vertebrates. They share the same yeast orthologue, Grn1p, but only GNL3L can rescue the growth-deficient phenotype in Grn1-null yeasts. To determine the unique function of GNL3L, we identified estrogen-related receptor gamma (ERRgamma) as a GNL3L-specific binding protein. GNL3L and ERRgamma are coexpressed in the eye, kidney and muscle, and co-reside in the nucleoplasm. The interaction between GNL3L and ERRgamma requires the intermediate domain of GNL3L and the AF2-domain of ERRgamma. Gain-of- and loss-of-function experiments show that GNL3L can inhibit the transcriptional activities of ERR genes in a cell-based reporter system, which does not require the nucleolar localization of GNL3L. We further demonstrate that GNL3L is able to reduce the steroid receptor coactivator (SRC) binding and the SRC-mediated transcriptional coactivation of ERRgamma. This work reveals a novel mechanism that negatively regulates the transcriptional function of ERRgamma by GNL3L through coactivator competition.
Figures





Similar articles
-
The homologous putative GTPases Grn1p from fission yeast and the human GNL3L are required for growth and play a role in processing of nucleolar pre-rRNA.Mol Biol Cell. 2006 Jan;17(1):460-74. doi: 10.1091/mbc.e05-09-0848. Epub 2005 Oct 26. Mol Biol Cell. 2006. PMID: 16251348 Free PMC article.
-
A novel lysine-rich domain and GTP binding motifs regulate the nucleolar retention of human guanine nucleotide binding protein, GNL3L.J Mol Biol. 2006 Dec 8;364(4):637-54. doi: 10.1016/j.jmb.2006.09.007. Epub 2006 Sep 8. J Mol Biol. 2006. PMID: 17034816
-
Interplay between estrogen-related receptor alpha (ERRalpha) and gamma (ERRgamma) on the regulation of ERRalpha gene expression.Mol Cell Endocrinol. 2007 Jan 29;264(1-2):128-41. doi: 10.1016/j.mce.2006.11.002. Epub 2006 Dec 8. Mol Cell Endocrinol. 2007. PMID: 17157980 Free PMC article.
-
Nucleolar modulation of TRF1: a dynamic way to regulate telomere and cell cycle by nucleostemin and GNL3L.Cell Cycle. 2009 Sep 15;8(18):2912-6. doi: 10.4161/cc.8.18.9543. Epub 2009 Sep 16. Cell Cycle. 2009. PMID: 19713769 Free PMC article. Review.
-
Nucleostemin: a latecomer with new tricks.Int J Biochem Cell Biol. 2009 Nov;41(11):2122-4. doi: 10.1016/j.biocel.2009.05.020. Epub 2009 Jun 6. Int J Biochem Cell Biol. 2009. PMID: 19501670 Free PMC article. Review.
Cited by
-
Maintenance of tumor initiating cells of defined genetic composition by nucleostemin.Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):20388-93. doi: 10.1073/pnas.1015171108. Epub 2011 Jul 5. Proc Natl Acad Sci U S A. 2011. PMID: 21730156 Free PMC article.
-
Leptin and insulin up-regulate miR-4443 to suppress NCOA1 and TRAF4, and decrease the invasiveness of human colon cancer cells.BMC Cancer. 2016 Nov 14;16(1):882. doi: 10.1186/s12885-016-2938-1. BMC Cancer. 2016. PMID: 27842582 Free PMC article.
-
On the Cutting Edge of Oral Cancer Prevention: Finding Risk-Predictive Markers in Precancerous Lesions by Longitudinal Studies.Cells. 2022 Mar 18;11(6):1033. doi: 10.3390/cells11061033. Cells. 2022. PMID: 35326482 Free PMC article. Review.
-
Multi-Omics Analysis of GNL3L Expression, Prognosis, and Immune Value in Pan-Cancer.Cancers (Basel). 2022 Sep 22;14(19):4595. doi: 10.3390/cancers14194595. Cancers (Basel). 2022. PMID: 36230520 Free PMC article.
-
GNL3L stabilizes the TRF1 complex and promotes mitotic transition.J Cell Biol. 2009 Jun 1;185(5):827-39. doi: 10.1083/jcb.200812121. J Cell Biol. 2009. PMID: 19487455 Free PMC article.
References
-
- Ariazi EA, Jordan VC. Estrogen-related receptors as emerging targets in cancer and metabolic disorders. Curr Top Med Chem. 2006;6:203–215. - PubMed
-
- Baddoo M, Hill K, Wilkinson R, Gaupp D, Hughes C, Kopen GC, Phinney DG. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem. 2003;89:1235–1249. - PubMed
-
- Bonnelye E, Vanacker JM, Spruyt N, Alric S, Fournier B, Desbiens X, Laudet V. Expression of the estrogen-related receptor 1 (ERR-1) orphan receptor during mouse development. Mech Dev. 1997;65:71–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

