Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals
- PMID: 17623783
- PMCID: PMC1924541
- DOI: 10.1073/pnas.0700531104
Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals
Abstract
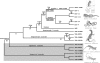
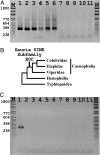
Poxviruses (Poxviridae) are a family of double-stranded DNA viruses with no RNA stage. Members of the genus Orthopoxvirus (OPV) are highly invasive and virulent. It was recently shown that the taterapox virus (TATV) from a West African rodent is the sister of camelpox virus and therefore belongs to the clade closest to the variola virus (VARV), the etiological agent of smallpox. Although these OPVs are among the most dreaded pathogens on Earth, our current knowledge of their genomes, their origins, and their possible hosts is still very limited. Here, we report the horizontal transfer of a retroposon (known only from reptilian genomes) to the TATV genome. After isolating and analyzing different subfamilies of short interspersed elements (SINEs) from lizards and snakes, we identified a highly poisonous snake (Echis ocellatus) from West Africa as the closest species from which the SINE sequence discovered in the TATV genome (TATV-SINE) was transferred to the virus. We discovered direct repeats derived from the virus flanking the TATV-SINE, and the absence of any snake-derived DNA flanking the SINE. These data provide strong evidence that the TATV-SINE was actually transferred within the snake to the viral genome by retrotransposition and not by any horizontal transfer at the DNA level. We propose that the snake is another host for TATV, suggesting that VARV-related epidemiologically relevant viruses may have derived from our cold-blooded ancestors and that poxviruses are possible vectors for horizontal transfer of retroposons from reptiles to mammals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

