Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs)
- PMID: 17623809
- PMCID: PMC1933517
- DOI: 10.1101/gr.6320607
Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs)
Abstract
Exonization of retroposed mobile elements, a process whereby new exons are generated following changes in non-protein-coding regions of a gene, is thought to have great potential for generating proteins with novel domains. Our previous analysis of primate-specific Alu-short interspersed elements (SINEs) showed, however, that during their 60 million years of evolution, SINE exonizations occurred in some primates, only to be lost again in some of the descendent lineages. This dynamic gain and loss makes it difficult to ascertain the contribution of exonization to genomic novelty. It was speculated that Alu-SINEs are too young to reveal persistent protein exaptation. In the present study we examined older mobile elements, mammalian-wide interspersed repeats (MIRs) that underwent active retroposition prior to the placental mammalian radiation approximately 130 million years ago, to determine their contribution to protein-coding sequences. Of 107 potential cases of MIR exonizations in human, an analysis of splice sites substantiates a mechanism that benefits from 3' splice site selection in MIR sequences. We retraced in detail the evolution of five MIR elements that exonized at different times during mammalian evolution. Four of these are expressed as alternatively spliced transcripts; three in species throughout the mammalian phylogenetic tree and one solely in primates. The fifth is the first experimentally verified, constitutively expressed retroposed SINE element in mammals. This pattern of highly conserved, alternatively and constitutively spliced MIR sequences evinces the potential of exonized transposed elements to evolve beyond the transient state found in Alu-SINEs and persist as important parts of functional proteins.
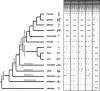
Figures



References
-
- Bejerano G., Lowe C.B., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Lowe C.B., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Rubin E.M., Kent W.J., Haussler D., Kent W.J., Haussler D., Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. - PubMed
-
- Bogaerts S., Vanlandschoot A., van Hengel J., van Roy F., Vanlandschoot A., van Hengel J., van Roy F., van Hengel J., van Roy F., van Roy F. Nuclear translocation of αN-catenin by the novel zinc finger transcriptional repressor ZASC1. Exp. Cell Res. 2005;311:1–13. - PubMed
-
- Bridges C.B. Genes and chromosomes. Teaching Biol. 1936;Nov:17–23.
-
- Brosius J. Echoes from the past—Are we still in an RNP world? Cytogenet. Genome Res. 2005;110:8–24. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
