Human disease classification in the postgenomic era: a complex systems approach to human pathobiology
- PMID: 17625512
- PMCID: PMC1948102
- DOI: 10.1038/msb4100163
Human disease classification in the postgenomic era: a complex systems approach to human pathobiology
Abstract
Contemporary classification of human disease derives from observational correlation between pathological analysis and clinical syndromes. Characterizing disease in this way established a nosology that has served clinicians well to the current time, and depends on observational skills and simple laboratory tools to define the syndromic phenotype. Yet, this time-honored diagnostic strategy has significant shortcomings that reflect both a lack of sensitivity in identifying preclinical disease, and a lack of specificity in defining disease unequivocally. In this paper, we focus on the latter limitation, viewing it as a reflection both of the different clinical presentations of many diseases (variable phenotypic expression), and of the excessive reliance on Cartesian reductionism in establishing diagnoses. The purpose of this perspective is to provide a logical basis for a new approach to classifying human disease that uses conventional reductionism and incorporates the non-reductionist approach of systems biomedicine.
Figures
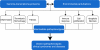
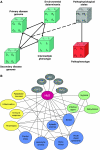
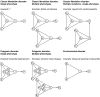


References
-
- Albert R (2005) Scale-free networks in cell biology. Journal Cell Sci 118: 4947–4957 - PubMed
-
- Albert R, Barabási AL (2002) Statistical mechanics of complex networks. Rev Mod Phys 74: 47–97
-
- Albert R, Jeong H, Barabasi AL (2000) Error and attack tolerance of complex networks. Nature 406: 378–382 - PubMed
-
- Barabási AL, Albert R (1999) Emergence of scaling in random networks. Science 286: 509–512 - PubMed
-
- Barabási AL, Oltvai Z (2004) Network biology: understanding the cell's functional organization. Nat Rev Genet 5: 101–113 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

