N-Glycan structure annotation of glycopeptides using a linearized glycan structure database (GlyDB)
- PMID: 17625816
- PMCID: PMC2557434
- DOI: 10.1021/pr070111y
N-Glycan structure annotation of glycopeptides using a linearized glycan structure database (GlyDB)
Abstract
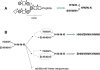
While glycoproteins are abundant in nature, and changes in glycosylation occur in cancer and other diseases, glycoprotein characterization remains a challenge due to the structural complexity of the biopolymers. This paper presents a general strategy, termed GlyDB, for glycan structure annotation of N-linked glycopeptides from tandem mass spectra in the LC-MS analysis of proteolytic digests of glycoproteins. The GlyDB approach takes advantage of low-energy collision-induced dissociation of N-linked glycopeptides that preferentially cleaves the glycosidic bonds while the peptide backbone remains intact. A theoretical glycan structure database derived from biosynthetic rules for N-linked glycans was constructed employing a novel representation of branched glycan structures consisting of multiple linear sequences. The commonly used peptide identification program, Sequest, could then be utilized to assign experimental tandem mass spectra to individual glycoforms. Analysis of synthetic glycopeptides and well-characterized glycoproteins demonstrate that the GlyDB approach can be a useful tool for annotation of glycan structures and for selection of a limited number of potential glycan structure candidates for targeted validation.
Figures


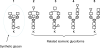
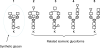


Similar articles
-
Characterization of intact N- and O-linked glycopeptides using higher energy collisional dissociation.Anal Biochem. 2014 May 1;452:96-102. doi: 10.1016/j.ab.2014.01.003. Epub 2014 Jan 15. Anal Biochem. 2014. PMID: 24440233 Free PMC article.
-
Automated Glycan Sequencing from Tandem Mass Spectra of N-Linked Glycopeptides.Anal Chem. 2016 Jun 7;88(11):5725-32. doi: 10.1021/acs.analchem.5b04858. Epub 2016 May 23. Anal Chem. 2016. PMID: 27111718 Free PMC article.
-
GlycoMaster DB: software to assist the automated identification of N-linked glycopeptides by tandem mass spectrometry.J Proteome Res. 2014 Sep 5;13(9):3881-95. doi: 10.1021/pr401115y. Epub 2014 Aug 25. J Proteome Res. 2014. PMID: 25113421
-
Analysis of glycoprotein-derived glycopeptides.EXS. 2000;88:159-86. doi: 10.1007/978-3-0348-8458-7_11. EXS. 2000. PMID: 10803378 Review.
-
Glycoproteomics based on tandem mass spectrometry of glycopeptides.J Chromatogr B Analyt Technol Biomed Life Sci. 2007 Apr 15;849(1-2):115-28. doi: 10.1016/j.jchromb.2006.09.041. Epub 2006 Oct 17. J Chromatogr B Analyt Technol Biomed Life Sci. 2007. PMID: 17049937 Review.
Cited by
-
Software for automated interpretation of mass spectrometry data from glycans and glycopeptides.Analyst. 2013 May 21;138(10):2793-803. doi: 10.1039/c2an36042j. Analyst. 2013. PMID: 23293784 Free PMC article. Review.
-
SweetSEQer, simple de novo filtering and annotation of glycoconjugate mass spectra.Mol Cell Proteomics. 2013 Jun;12(6):1735-40. doi: 10.1074/mcp.O112.025940. Epub 2013 Feb 26. Mol Cell Proteomics. 2013. PMID: 23443135 Free PMC article.
-
Generation of asparagine-linked glycan structure databases and their use.J Am Soc Mass Spectrom. 2009 Sep;20(9):1739-42. doi: 10.1016/j.jasms.2009.05.012. Epub 2009 May 29. J Am Soc Mass Spectrom. 2009. PMID: 19556140
-
GlycoPep Detector: a tool for assigning mass spectrometry data of N-linked glycopeptides on the basis of their electron transfer dissociation spectra.Anal Chem. 2013 May 21;85(10):5023-32. doi: 10.1021/ac400287n. Epub 2013 Apr 29. Anal Chem. 2013. PMID: 23510108 Free PMC article.
-
Exploring site-specific N-glycosylation microheterogeneity of haptoglobin using glycopeptide CID tandem mass spectra and glycan database search.J Proteome Res. 2013 Aug 2;12(8):3652-66. doi: 10.1021/pr400196s. Epub 2013 Jul 22. J Proteome Res. 2013. PMID: 23829323 Free PMC article.
References
-
- Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nat Methods. 2005;2(11):817–24. - PubMed
-
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473(1):4–8. - PubMed
-
- Woods RJ, Edge CJ, Dwek RA. Protein surface oligosaccharides and protein function. Nat Struct Biol. 1994;1(8):499–501. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

