Arterivirus subgenomic mRNA synthesis and virion biogenesis depend on the multifunctional nsp1 autoprotease
- PMID: 17626105
- PMCID: PMC2045461
- DOI: 10.1128/JVI.00683-07
Arterivirus subgenomic mRNA synthesis and virion biogenesis depend on the multifunctional nsp1 autoprotease
Abstract
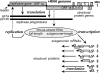
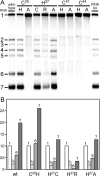
Many groups of plus-stranded RNA viruses produce additional, subgenomic mRNAs to regulate the expression of part of their genome. Arteriviruses and coronaviruses (order Nidovirales) are unique among plus-stranded RNA viruses for using a mechanism of discontinuous RNA synthesis to produce a nested set of 5'- and 3'-coterminal subgenomic mRNAs, which serve to express the viral structural protein genes. The discontinuous step presumably occurs during minus-strand synthesis and joins noncontiguous sequences copied from the 3'- and 5'-proximal domains of the genomic template. Nidovirus genome amplification ("replication") and subgenomic mRNA synthesis ("transcription") are driven by 13 to 16 nonstructural proteins (nsp's), generated by autocatalytic processing of two large "replicase" polyproteins. Previously, using a replicon system, the N-terminal nsp1 replicase subunit of the arterivirus equine arteritis virus (EAV) was found to be dispensable for replication but crucial for transcription. Using reverse genetics, we have now addressed the role of nsp1 against the background of the complete EAV life cycle. Mutagenesis revealed that nsp1 is in fact a multifunctional regulatory protein. Its papain-like autoprotease domain releases nsp1 from the replicase polyproteins, a cleavage essential for viral RNA synthesis. Several mutations in the putative N-terminal zinc finger domain of nsp1 selectively abolished transcription, while replication was either not affected or even increased. Other nsp1 mutations did not significantly affect either replication or transcription but still dramatically reduced the production of infectious progeny. Thus, nsp1 is involved in at least three consecutive key processes in the EAV life cycle: replicase polyprotein processing, transcription, and virion biogenesis.
Figures




Similar articles
-
Arterivirus Nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs.PLoS Pathog. 2010 Feb 19;6(2):e1000772. doi: 10.1371/journal.ppat.1000772. PLoS Pathog. 2010. PMID: 20174607 Free PMC article.
-
The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis.J Virol. 2000 Jun;74(11):5213-23. doi: 10.1128/jvi.74.11.5213-5223.2000. J Virol. 2000. PMID: 10799597 Free PMC article.
-
A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus.Proc Natl Acad Sci U S A. 2001 Feb 13;98(4):1889-94. doi: 10.1073/pnas.98.4.1889. Epub 2001 Feb 6. Proc Natl Acad Sci U S A. 2001. PMID: 11172046 Free PMC article.
-
Nidovirus RNA polymerases: Complex enzymes handling exceptional RNA genomes.Virus Res. 2017 Apr 15;234:58-73. doi: 10.1016/j.virusres.2017.01.023. Epub 2017 Feb 6. Virus Res. 2017. PMID: 28174054 Free PMC article. Review.
-
Biogenesis and architecture of arterivirus replication organelles.Virus Res. 2016 Jul 15;220:70-90. doi: 10.1016/j.virusres.2016.04.001. Epub 2016 Apr 9. Virus Res. 2016. PMID: 27071852 Free PMC article. Review.
Cited by
-
Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design.Nat Rev Mol Cell Biol. 2022 Jan;23(1):21-39. doi: 10.1038/s41580-021-00432-z. Epub 2021 Nov 25. Nat Rev Mol Cell Biol. 2022. PMID: 34824452 Free PMC article. Review.
-
Targeting Swine Leukocyte Antigen Class I Molecules for Proteasomal Degradation by the nsp1α Replicase Protein of the Chinese Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus Strain JXwn06.J Virol. 2015 Oct 21;90(2):682-93. doi: 10.1128/JVI.02307-15. Print 2016 Jan 15. J Virol. 2015. PMID: 26491168 Free PMC article.
-
The zinc-finger domain was essential for porcine reproductive and respiratory syndrome virus nonstructural protein-1α to inhibit the production of interferon-β.J Interferon Cytokine Res. 2013 Jun;33(6):328-34. doi: 10.1089/jir.2012.0100. Epub 2013 Feb 21. J Interferon Cytokine Res. 2013. PMID: 23428052 Free PMC article.
-
Equine arteritis virus.Vet Microbiol. 2013 Nov 29;167(1-2):93-122. doi: 10.1016/j.vetmic.2013.06.015. Epub 2013 Jul 3. Vet Microbiol. 2013. PMID: 23891306 Free PMC article. Review.
-
Modulation of innate immune signaling by nonstructural protein 1 (nsp1) in the family Arteriviridae.Virus Res. 2014 Dec 19;194:100-9. doi: 10.1016/j.virusres.2014.09.007. Epub 2014 Sep 28. Virus Res. 2014. PMID: 25262851 Free PMC article. Review.
References
-
- Baker, S. C., N. La Monica, C. K. Shieh, and M. M. C. Lai. 1990. Murine coronavirus gene 1 polyprotein contains an autoproteolytic activity. Adv. Exp. Biol. Med. 276:283-289. - PubMed
-
- Ball, L. A. 2001. Replication strategies of RNA viruses, 105-118. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott, Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

