Down-regulation of SFRP1 as a putative tumor suppressor gene can contribute to human hepatocellular carcinoma
- PMID: 17626620
- PMCID: PMC1940018
- DOI: 10.1186/1471-2407-7-126
Down-regulation of SFRP1 as a putative tumor suppressor gene can contribute to human hepatocellular carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is one of the most common cancers in the world. SFRP1 (the secreted frizzled-related protein 1), a putative tumor suppressor gene mapped onto chromosome 8p12-p11.1, the frequent loss of heterozygosity (LOH) region in human HCC, encodes a Wingless-type (Wnt) signaling antagonist and is frequently inactivated by promoter methylation in many human cancers. However, whether the down-regulation of SFRP1 can contribute to hepatocarcinogenesis still remains unclear.
Methods: We investigated the expression of SFRP1 through real time RT-PCR and immunohistochemistry staining. The cell growth and colony formation were observed as the overexpression and knockdown of SFRP1. The DNA methylation status within SFRP1 promoter was analyzed through methylation-specific PCR or bisulphate-treated DNA sequencing assays. Loss of heterozygosity was here detected with microsatellite markers.
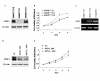
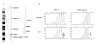
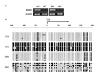
Results: SFRP1 was significantly down-regulated in 76.1% (35/46) HCC specimens at mRNA level and in 30% (30/100) HCCs indicated by immunohistochemistry staining, as compared to adjacent non-cancerous livers. The overexpression of SFRP1 can significantly inhibit the cell growth and colony formation of YY-8103, SMMC7721, and Hep3B cells. The RNA interference against the constitutional SFRP1 in the offspring SMMC7721 cells, which were stably transfected by ectopic SFRP1, can markedly promote cell growth of these cells. LOH of both microsatellite markers D8S532 and D8SAC016868 flanking the gene locus was found in 13% (6 of 46 HCCs) and 6.5% (3 of 46 HCCs) of the informative cases, respectively, where 5 of 8 HCC specimens with LOH showed the down-regulation of SFRP1. DNA hypermethylation within SFRP1 promoter was identified in two of three HCC specimens without SFRP1 expression. Moreover, the DNA methylation of SFRP1 promoter was significantly reduced, along with the re-expression of the gene, in those HCC cell lines, Bel7404, QGY7701, and MHCC-H, as treated by DAC.
Conclusion: Our data suggested that the down-regulation of SFRP1 as a candidate tumor suppressor gene, triggered by the epigenetic and/or genetic events, could contribute to the oncogenesis of HCC.
Figures



Similar articles
-
Promoter methylation of the secreted frizzled-related protein 1 gene SFRP1 is frequent in hepatocellular carcinoma.Cancer. 2006 Aug 1;107(3):579-90. doi: 10.1002/cncr.22023. Cancer. 2006. PMID: 16795071
-
Epigenetic inactivation of the canonical Wnt antagonist secreted frizzled-related protein 1 in hepatocellular carcinoma cells.Neoplasma. 2012;59(3):326-32. doi: 10.4149/neo_2012_042. Neoplasma. 2012. PMID: 22296502
-
Inhibition mechanism of a newly cloned candidate tumor suppressor gene JST during hepatocarcinogenesis and its abnormal expression in human hepatocellular carcinoma from Qidong liver cancer risk area, China.Hepatogastroenterology. 2004 Mar-Apr;51(56):515-25. Hepatogastroenterology. 2004. PMID: 15086194
-
Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma.World J Gastroenterol. 2015 May 28;21(20):6317-28. doi: 10.3748/wjg.v21.i20.6317. World J Gastroenterol. 2015. PMID: 26034368 Free PMC article. Review.
-
DNA methylation in hepatocellular carcinoma.World J Gastroenterol. 2008 Mar 21;14(11):1741-8. doi: 10.3748/wjg.14.1741. World J Gastroenterol. 2008. PMID: 18350605 Free PMC article. Review.
Cited by
-
Downregulation of SFRP1 is a protumorigenic event in hepatoblastoma and correlates with beta-catenin mutations.J Cancer Res Clin Oncol. 2020 May;146(5):1153-1167. doi: 10.1007/s00432-020-03182-1. Epub 2020 Mar 18. J Cancer Res Clin Oncol. 2020. PMID: 32189106 Free PMC article.
-
Predicting hepatocellular carcinoma development for cirrhosis patients via methylation detection of heparocarcinogenesis-related genes.J Cancer. 2018 Jun 4;9(12):2203-2210. doi: 10.7150/jca.24024. eCollection 2018. J Cancer. 2018. PMID: 29937940 Free PMC article.
-
Dependence receptor UNC5D mediates nerve growth factor depletion-induced neuroblastoma regression.J Clin Invest. 2013 Jul;123(7):2935-47. doi: 10.1172/JCI65988. Epub 2013 Jun 17. J Clin Invest. 2013. PMID: 23778138 Free PMC article.
-
Using PDX animal models to identify and stratify adenoid cystic carcinoma patients presenting an enhanced response to HDAC inhibitors.Am J Cancer Res. 2023 Jan 15;13(1):143-160. eCollection 2023. Am J Cancer Res. 2023. PMID: 36777521 Free PMC article.
-
Deregulation of Frizzled Receptors in Hepatocellular Carcinoma.Int J Mol Sci. 2018 Jan 21;19(1):313. doi: 10.3390/ijms19010313. Int J Mol Sci. 2018. PMID: 29361730 Free PMC article. Review.
References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
-
- Wang JS, Huang T, Su J, Liang F, Wei Z, Liang Y, Luo H, Kuang SY, Qian GS, Sun G, He X, Kensler TW, Groopman JD. Hepatocellular carcinoma and aflatoxin exposure in Zhuqing Village, Fusui County, People's Republic of China. Cancer Epidemiol Biomarkers Prev. 2001;10:143–146. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

