Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana
- PMID: 17626786
- PMCID: PMC1914337
- DOI: 10.1073/pnas.0705323104
Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana
Abstract
We used hybridization to the ATH1 gene expression array to interrogate genomic DNA diversity in 23 wild strains (accessions) of Arabidopsis thaliana (arabidopsis), in comparison with the reference strain Columbia (Col). At <1% false discovery rate, we detected 77,420 single-feature polymorphisms (SFPs) with distinct patterns of variation across the genome. Total and pair-wise diversity was higher near the centromeres and the heterochromatic knob region, but overall diversity was positively correlated with recombination rate (R(2) = 3.1%). The difference between total and pair-wise SFP diversity is a relative measure contrasting diversifying or frequency-dependent selection, similar to Tajima's D, and can be calibrated by the empirical genome-wide distribution. Each unique locus, centered on a gene, has a diversity and selection score that suggest a relative role in past evolutionary processes. Homologs of disease resistance (R) genes include members with especially high levels of diversity often showing frequency-dependent selection and occasionally evidence of a past selective sweep. Receptor-like and S-locus proteins also contained members with elevated levels of diversity and signatures of selection, whereas other gene families, bHLH, F-box, and RING finger proteins, showed more typical levels of diversity. SFPs identified with the gene expression array also provide an empirical hybridization polymorphism background for studies of gene expression polymorphism and are available through the genome browser http://signal.salk.edu/cgi-bin/AtSFP.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

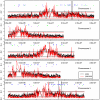

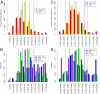
Comment in
-
An Ultra-High-Density, Transcript-Based, Genetic Map of Lettuce.G3 (Bethesda). 2013 Apr 9;3(4):617-631. doi: 10.1534/g3.112.004929. G3 (Bethesda). 2013. PMID: 23550116 Free PMC article.
References
-
- Yoshida K, Kamiya T, Kawabe A, Miyashita NT. Genes Genet Syst. 2003;78:11–21. - PubMed
-
- Sharbel TF, Haubold B, Mitchell-Olds T. Mol Ecol. 2000;9:2109–2118. - PubMed
-
- Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloof JN, Noyes T, Oefner PJ, et al. Nat Genet. 2002;30:190–193. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

