A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway
- PMID: 17626790
- PMCID: PMC1948855
- DOI: 10.1101/gad.1565607
A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway
Abstract
Binding of microRNA (miRNA) to mRNA within the RNA-induced silencing complex (RISC) leads to either translational inhibition or to destruction of the target mRNA. Both of these functions are executed by Argonaute 2 (Ago2). Using hematopoiesis in mice as a model system to study the physiological function of Ago2 in vivo, we found that Ago2 controls early development of lymphoid and erythroid cells. We show that the unique and defining feature of Ago2, the Slicer endonuclease activity, is dispensable for hematopoiesis. Instead, we identified Ago2 as a key regulator of miRNA homeostasis. Deficiency in Ago2 impairs miRNA biogenesis from precursor-miRNAs followed by a reduction in miRNA expression levels. Collectively, our data identify Ago2 as a highly specialized member of the Argonaute family with an essential nonredundant Slicer-independent function within the mammalian miRNA pathway.
Figures
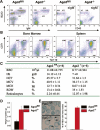
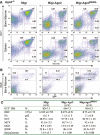
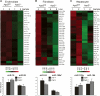
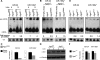
Comment in
-
Life beyond cleavage: the case of Ago2 and hematopoiesis.Genes Dev. 2007 Aug 15;21(16):1983-8. doi: 10.1101/gad.1591407. Genes Dev. 2007. PMID: 17699747 Review. No abstract available.
References
-
- Chen C.Z., Li L., Lodish H.F., Bartel D.P., Li L., Lodish H.F., Bartel D.P., Lodish H.F., Bartel D.P., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. - PubMed
-
- Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R., Norman J., Cooch N., Nishikura K., Shiekhattar R., Cooch N., Nishikura K., Shiekhattar R., Nishikura K., Shiekhattar R., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. - PMC - PubMed
-
- Cobb B.S., Nesterova T.B., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Nesterova T.B., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Brockdorff N., Fisher A.G., Smale S.T., Fisher A.G., Smale S.T., Smale S.T., et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 2005;201:1367–1373. - PMC - PubMed
-
- Farley F.W., Soriano P., Steffen L.S., Dymecki S.M., Soriano P., Steffen L.S., Dymecki S.M., Steffen L.S., Dymecki S.M., Dymecki S.M. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
