Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma
- PMID: 17627275
- PMCID: PMC1949000
- DOI: 10.1038/sj.emboj.7601794
Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma
Abstract
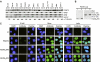
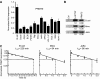
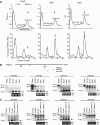
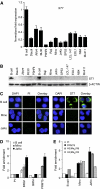
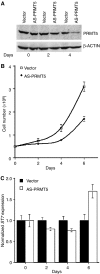
Protein arginine methyltransferase PRMT5 interacts with human SWI/SNF complexes and methylates histones H3R8 and H4R3. To elucidate the role of PRMT5 in human cancer, we analyzed PRMT5 expression in normal human B lymphocytes and a panel of lymphoid cancer cell lines as well as mantle cell lymphoma (MCL) clinical samples. We show that PRMT5 protein levels are elevated in all cancer cells, including clinical samples examined despite its low rate of transcription and messenger RNA stability. Remarkably, polysome profiling revealed that PRMT5 mRNA is translated more efficiently in Mino and JeKo MCL cells than in normal B cells, and that decreased miR-92b and miR-96 expression augments PRMT5 translation. Consequently, global methylation of H3R8 and H4R3 is increased and is accompanied by repression of suppressor of tumorigenecity 7 (ST7) in lymphoid cancer cells. Furthermore, knockdown of PRMT5 expression reduces proliferation of transformed JeKo and Raji cells. Thus, our studies indicate that aberrant expression of PRMT5 leads to altered epigenetic modification of chromatin, which in turn impacts transcriptional performance of anti-cancer genes and growth of transformed lymphoid cells.
Figures



References
-
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzaride T, Surani MA (2006) Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol 8: 623–630 - PubMed
-
- Bedford MT, Richard S (2005) Arginine methylation an emerging regulator of protein function. Mol Cell 18: 263–272 - PubMed
-
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in polycomb -group silencing. Science 298: 1039–1043 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

