Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration
- PMID: 17627281
- PMCID: PMC1949006
- DOI: 10.1038/sj.emboj.7601787
Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration
Abstract
Protection from NO gas, a toxic byproduct of anaerobic respiration in Pseudomonas aeruginosa, is mediated by nitric oxide (NO) reductase (NOR), the norCB gene product. Nevertheless, a norCB mutant that accumulated approximately 13.6 microM NO paradoxically survived anaerobic growth. Transcription of genes encoding nitrate and nitrite reductases, the enzymes responsible for NO production, was reduced >50- and 2.5-fold in the norCB mutant. This was due, in part, to a predicted compromise of the [4Fe-4S](2+) cluster in the anaerobic regulator ANR by physiological NO levels, resulting in an inability to bind to its cognate promoter DNA sequences. Remarkably, two O(2)-dependent dioxygenases, homogentisate-1,2-dioxygenase (HmgA) and 4-hydroxyphenylpyruvate dioxygenase (Hpd), were derepressed in the norCB mutant. Electron paramagnetic resonance studies showed that HmgA and Hpd bound NO avidly, and helped protect the norCB mutant in anaerobic biofilms. These data suggest that protection of a P. aeruginosa norCB mutant against anaerobic NO toxicity occurs by both control of NO supply and reassignment of metabolic enzymes to the task of NO sequestration.
Figures
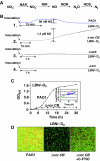





 ). In contrast, genes that are anaerobically repressed, including hmgA and hpd, become derepressed, and function as NO scavengers; (4) because ANR is required for transcription of dnr, DNR is not produced; (5) thus, narK1, nirS, nirQ and oprE transcripts are either absent or extremely low (denoted by red X); (6) ANR, and perhaps DNR, denoted as a ‘?', however, loses it activity as a repressor. Thus, genes that would be repressed anaerobically, such as hmgA and hpd, are now derepressed (denoted by green circle). Because of the inherent NO-binding properties of HmgA and Hpd (refer to Figure 5A and B), elevated levels of these enzymes served to help protect the anaerobic norCB mutant against NO-mediated toxicity. OM, outer membrane; PP, periplasmic space; IM, inner membrane; NarK1/2, putative NO2− extrusion pump; CytC (red), reduced cytochrome c; Azurin, periplasmic protein donating electrons to NIR; e−, electron flow; NADH, electron donor for NAR.
). In contrast, genes that are anaerobically repressed, including hmgA and hpd, become derepressed, and function as NO scavengers; (4) because ANR is required for transcription of dnr, DNR is not produced; (5) thus, narK1, nirS, nirQ and oprE transcripts are either absent or extremely low (denoted by red X); (6) ANR, and perhaps DNR, denoted as a ‘?', however, loses it activity as a repressor. Thus, genes that would be repressed anaerobically, such as hmgA and hpd, are now derepressed (denoted by green circle). Because of the inherent NO-binding properties of HmgA and Hpd (refer to Figure 5A and B), elevated levels of these enzymes served to help protect the anaerobic norCB mutant against NO-mediated toxicity. OM, outer membrane; PP, periplasmic space; IM, inner membrane; NarK1/2, putative NO2− extrusion pump; CytC (red), reduced cytochrome c; Azurin, periplasmic protein donating electrons to NIR; e−, electron flow; NADH, electron donor for NAR.Similar articles
-
Functional roles of norCB in Pseudomonas aeruginosa ATCC 9027 under aerobic conditions.J Basic Microbiol. 2019 Nov;59(11):1154-1162. doi: 10.1002/jobm.201900267. Epub 2019 Sep 25. J Basic Microbiol. 2019. PMID: 31553498
-
Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis.Dev Cell. 2002 Oct;3(4):593-603. doi: 10.1016/s1534-5807(02)00295-2. Dev Cell. 2002. PMID: 12408810
-
Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets.Adv Drug Deliv Rev. 2002 Dec 5;54(11):1425-43. doi: 10.1016/s0169-409x(02)00152-7. Adv Drug Deliv Rev. 2002. PMID: 12458153 Review.
-
Catalase (KatA) plays a role in protection against anaerobic nitric oxide in Pseudomonas aeruginosa.PLoS One. 2014 Mar 24;9(3):e91813. doi: 10.1371/journal.pone.0091813. eCollection 2014. PLoS One. 2014. PMID: 24663218 Free PMC article.
-
Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways.Trends Microbiol. 2009 Mar;17(3):130-8. doi: 10.1016/j.tim.2008.12.003. Epub 2009 Feb 21. Trends Microbiol. 2009. PMID: 19231190 Review.
Cited by
-
Sodium nitrite blocks the activity of aminoglycosides against Pseudomonas aeruginosa biofilms.Antimicrob Agents Chemother. 2015;59(6):3329-34. doi: 10.1128/AAC.00546-15. Epub 2015 Mar 23. Antimicrob Agents Chemother. 2015. PMID: 25801569 Free PMC article.
-
Nitrous oxide production and consumption: regulation of gene expression by gas-sensitive transcription factors.Philos Trans R Soc Lond B Biol Sci. 2012 May 5;367(1593):1213-25. doi: 10.1098/rstb.2011.0309. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22451107 Free PMC article. Review.
-
The histone-like protein AlgP regulon is distinct in mucoid and nonmucoid Pseudomonas aeruginosa and does not include alginate biosynthesis genes.Microbiology (Reading). 2020 Sep;166(9):861-866. doi: 10.1099/mic.0.000923. Microbiology (Reading). 2020. PMID: 32634088 Free PMC article.
-
A Commensal Streptococcus Dysregulates the Pseudomonas aeruginosa Nitrosative Stress Response.Front Cell Infect Microbiol. 2022 May 10;12:817336. doi: 10.3389/fcimb.2022.817336. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35619650 Free PMC article.
-
Acquisition and role of molybdate in Pseudomonas aeruginosa.Appl Environ Microbiol. 2014 Nov;80(21):6843-52. doi: 10.1128/AEM.02465-14. Epub 2014 Aug 29. Appl Environ Microbiol. 2014. PMID: 25172858 Free PMC article.
References
-
- Arai H, Igarashi Y, Kodama T (1994) Structure and ANR-dependent transcription of the nir genes for denitrification from Pseudomonas aeruginosa. Biosci Biotechnol Biochem 58: 1286–1291 - PubMed
-
- Arai H, Igarashi Y, Kodama T (1995) Expression of the nir and nor genes for denitrification of Pseudomonas aeruginosa requires a novel CRP/FNR-related transcriptional regulator, DNR, in addition to ANR. FEBS Lett 371: 73–76 - PubMed
-
- Arai H, Kodama T, Igarashi Y (1997) Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol Microbiol 25: 1141–1148 - PubMed
-
- Arai H, Mizutani M, Igarashi Y (2003) Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 149: 29–36 - PubMed
-
- Arciero DM, Lipscomb JD (1986) Binding of 17O-labeled substrate and inhibitors to protocatechuate 4,5-dioxygenase-nitrosyl complex. Evidence for direct substrate binding to the active site Fe2+ of extradiol dioxygenases. J Biol Chem 261: 2170–2178 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

