Study of the mode of action of a polygalacturonase from the phytopathogen Burkholderia cepacia
- PMID: 17627609
- PMCID: PMC2049012
- DOI: 10.1042/BJ20061833
Study of the mode of action of a polygalacturonase from the phytopathogen Burkholderia cepacia
Abstract
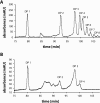
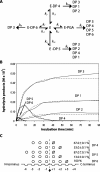
We have recently isolated and heterologously expressed BcPeh28A, an endopolygalacturonase from the phytopathogenic Gram-negative bacterium Burkholderia cepacia. Endopolygalacturonases belong to glycoside hydrolase family 28 and are responsible for the hydrolysis of the non-esterified regions of pectins. The mode of action of BcPeh28A on different substrates has been investigated and its enzymatic mechanism elucidated. The hydrolysis of polygalacturonate indicates that BcPeh28A is a non-processive enzyme that releases oligomers with chain lengths ranging from two to eight. By inspection of product progression curves, a kinetic model has been generated and extensively tested. It has been used to derive the kinetic parameters that describe the time course of the formation of six predominant products. Moreover, an investigation of the enzymatic activity on shorter substrates that differ in their overall length and methylation patterns sheds light on the architecture of the BcPeh28A active site. Specifically the tolerance of individual sites towards methylated saccharide units was rationalized on the basis of the hydrolysis of hexagalacturonides with different methylation patterns.
Figures




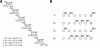


Similar articles
-
Insight into the structure of an endopolygalacturonase from the phytopathogen Burkholderia cepacia: a biochemical and computational study.Biochimie. 2010 Oct;92(10):1445-53. doi: 10.1016/j.biochi.2010.07.007. Epub 2010 Jul 15. Biochimie. 2010. PMID: 20637827
-
Study of the mode of action of endopolygalacturonase from Fusarium moniliforme.Biochim Biophys Acta. 2001 Jun 15;1526(3):301-9. doi: 10.1016/s0304-4165(01)00141-6. Biochim Biophys Acta. 2001. PMID: 11410340
-
Isolation, heterologous expression and characterization of an endo-polygalacturonase produced by the phytopathogen Burkholderia cepacia.Protein Expr Purif. 2007 Aug;54(2):300-8. doi: 10.1016/j.pep.2007.03.019. Epub 2007 Apr 10. Protein Expr Purif. 2007. PMID: 17493828
-
Hydrolysis of pectins with different degrees and patterns of methylation by the endopolygalacturonase of Fusarium moniliforme.Biochim Biophys Acta. 2002 Apr 1;1596(1):83-94. doi: 10.1016/s0167-4838(02)00207-8. Biochim Biophys Acta. 2002. PMID: 11983424
-
Kinetic characterization of Aspergillus niger N400 endopolygalacturonases I, II and C.Eur J Biochem. 1999 Feb;259(3):577-85. doi: 10.1046/j.1432-1327.1999.00080.x. Eur J Biochem. 1999. PMID: 10092840
Cited by
-
Functional Classification and Characterization of the Fungal Glycoside Hydrolase 28 Protein Family.J Fungi (Basel). 2022 Feb 22;8(3):217. doi: 10.3390/jof8030217. J Fungi (Basel). 2022. PMID: 35330219 Free PMC article.
References
-
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., et al. Toward a systems approach to understanding plant cell walls. Science. 2003;306:2206–2211. - PubMed
-
- Willats W. G., McCartney L., Mackie W., Knox J. P. Pectin: cell biology and prospects for functional analysis. Plant Mol. Biol. 2001;47:9–27. - PubMed
-
- Ridley B. L., O'Neill M. A., Mohnen D. Pectins: structure, biosynthesis and oligogalacturonide-related signaling. Phytochemistry. 2001;57:929–967. - PubMed
-
- Prade R. A., Zhan D., Ayoubi P., Mort A. J. Pectins, pectinases and plant–microbe interactions. Biotechnol. Genet. Eng. Rev. 1999;16:361–391. - PubMed
-
- Collmer A., Keen N. T. The role of pectic enzymes in plant pathogenesis. Annu. Rev. Phytopathol. 1986;24:383–409.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

