Using two-component systems and other bacterial regulatory factors for the fabrication of synthetic genetic devices
- PMID: 17628156
- PMCID: PMC3052260
- DOI: 10.1016/S0076-6879(06)22025-1
Using two-component systems and other bacterial regulatory factors for the fabrication of synthetic genetic devices
Abstract
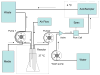
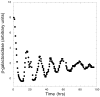
Synthetic biology is an emerging field in which the procedures and methods of engineering are extended living organisms, with the long-term goal of producing novel cell types that aid human society. For example, engineered cell types may sense a particular environment and express gene products that serve as an indicator of that environment or affect a change in that environment. While we are still some way from producing cells with significant practical applications, the immediate goals of synthetic biology are to develop a quantitative understanding of genetic circuitry and its interactions with the environment and to develop modular genetic circuitry derived from standard, interoperable parts that can be introduced into cells and result in some desired input/output function. Using an engineering approach, the input/output function of each modular element is characterized independently, providing a toolkit of elements that can be linked in different ways to provide various circuit topologies. The principle of modularity, yet largely unproven for biological systems, suggests that modules will function appropriately based on their design characteristics when combined into larger synthetic genetic devices. This modularity concept is similar to that used to develop large computer programs, where independent software modules can be independently developed and later combined into the final program. This chapter begins by pointing out the potential usefulness of two-component signal transduction systems for synthetic biology applications and describes our use of the Escherichia coli NRI/NRII (NtrC/NtrB) two-component system for the construction of a synthetic genetic oscillator and toggle switch for E. coli. Procedures for conducting measurements of oscillatory behavior and toggle switch behavior of these synthetic genetic devices are described. It then presents a brief overview of device fabrication strategy and tactics and presents a useful vector system for the construction of synthetic genetic modules and positioning these modules onto the bacterial chromosome in defined locations.
Figures


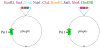


References
-
- Atkinson MR, Pattaramanon N, Ninfa AJ. Governor of the glnAp2 promoter of Escherichia coli. Mol Microbiol. 2002;46:1247–1257. - PubMed
-
- Atkinson MR, Savageau MA, Meyers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

