Novel exon combinations generated by alternative splicing of gene fragments mobilized by a CACTA transposon in Glycine max
- PMID: 17629935
- PMCID: PMC1947982
- DOI: 10.1186/1471-2229-7-38
Novel exon combinations generated by alternative splicing of gene fragments mobilized by a CACTA transposon in Glycine max
Abstract
Background: The recent discoveries of transposable elements carrying host gene fragments such as the Pack-MULEs (Mutator-like transposable elements) of maize (Zea mays), rice (Oryza sativa) and Arabidopsis thaliana, the Helitrons of maize and the Tgm-Express of soybeans, revealed a widespread genetic mechanism with the potential to rearrange genomes and create novel chimeric genes affecting genomic and proteomic diversity. Not much is known with regard to the mechanisms of gene fragment capture by those transposon elements or the expression of the captured host gene fragments. There is some evidence that chimeric transcripts can be assembled and exist in EST collections.
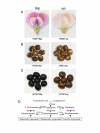
Results: We report results obtained from analysis of RT-PCR derived cDNAs of the Glycine max mutant flower color gene, wp, that contains a 5.7-kb transposon (Tgm-Express1) in Intron 2 of the flavanone 3-hydroxylase gene (F3H) and is composed of five unrelated host gene fragments. The collection of cDNAs derived from the wp allele represents a multiplicity of processed RNAs varying in length and sequence that includes some identical to the correctly processed mature F3H transcript with three properly spliced exons. Surprisingly, the five gene fragments carried by the Tgm-Express1 were processed through complex alternative splicing as additional exons of the wp transcript.
Conclusion: The gene fragments carried by the Tgm inverted repeat ends appear to be retained as functional exons/introns within the element. The spliceosomes then select indiscriminately the canonical intron splice sites from a pre-mRNA to assemble diverse chimeric transcripts from the exons contained in the wp allele. The multiplicity and randomness of these events provide some insights into the origin and mechanism of alternatively spliced genes.
Figures



Similar articles
-
The wp mutation of Glycine max carries a gene-fragment-rich transposon of the CACTA superfamily.Plant Cell. 2005 Oct;17(10):2619-32. doi: 10.1105/tpc.105.033506. Epub 2005 Sep 2. Plant Cell. 2005. PMID: 16141454 Free PMC article.
-
A putative autonomous 20.5 kb-CACTA transposon insertion in an F3'H allele identifies a new CACTA transposon subfamily in Glycine max.BMC Plant Biol. 2008 Dec 2;8:124. doi: 10.1186/1471-2229-8-124. BMC Plant Biol. 2008. PMID: 19055742 Free PMC article.
-
Suppression of an atypically spliced rice CACTA transposon transcript in transgenic plants.Genetics. 2005 Apr;169(4):2383-7. doi: 10.1534/genetics.104.037358. Epub 2005 Jan 31. Genetics. 2005. PMID: 15687269 Free PMC article.
-
Pack-MULE transposable elements mediate gene evolution in plants.Nature. 2004 Sep 30;431(7008):569-73. doi: 10.1038/nature02953. Nature. 2004. PMID: 15457261
-
Exonization of transposed elements: A challenge and opportunity for evolution.Biochimie. 2011 Nov;93(11):1928-34. doi: 10.1016/j.biochi.2011.07.014. Epub 2011 Jul 26. Biochimie. 2011. PMID: 21787833 Review.
Cited by
-
Genome-wide survey of ds exonization to enrich transcriptomes and proteomes in plants.Evol Bioinform Online. 2012;8:575-87. doi: 10.4137/EBO.S10324. Epub 2012 Oct 8. Evol Bioinform Online. 2012. PMID: 23091369 Free PMC article.
-
Editor's choice: Crop genome plasticity and its relevance to food and feed safety of genetically engineered breeding stacks.Plant Physiol. 2012 Dec;160(4):1842-53. doi: 10.1104/pp.112.204271. Epub 2012 Oct 11. Plant Physiol. 2012. PMID: 23060369 Free PMC article. Review. No abstract available.
-
Mobile DNA and evolution in the 21st century.Mob DNA. 2010 Jan 25;1(1):4. doi: 10.1186/1759-8753-1-4. Mob DNA. 2010. PMID: 20226073 Free PMC article.
-
Recent amplification and impact of MITEs on the genome of grapevine (Vitis vinifera L.).Genome Biol Evol. 2009 May 20;1:75-84. doi: 10.1093/gbe/evp009. Genome Biol Evol. 2009. PMID: 20333179 Free PMC article.
-
Androgenic-Induced Transposable Elements Dependent Sequence Variation in Barley.Int J Mol Sci. 2021 Jun 24;22(13):6783. doi: 10.3390/ijms22136783. Int J Mol Sci. 2021. PMID: 34202586 Free PMC article.
References
-
- Stephens PA, Nickell CD. Inheritance of pink flower color in soybean. Crop Sci. 1992;32:1131–1132.
-
- Stephens PA, Nickell CD, Vodkin LO. Pink flower color associated with increased protein and seed size in soybean. Crop Sci. 1993;33:1135–1137.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

