Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments
- PMID: 17630306
- PMCID: PMC2074903
- DOI: 10.1128/AEM.00643-07
Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments
Abstract
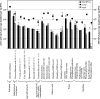
Dissimilatory nitrate reduction is catalyzed by a membrane-bound and a periplasmic nitrate reductase. We set up a real-time PCR assay to quantify these two enzymes, using the narG and napA genes, encoding the catalytic subunits of the two types of nitrate reductases, as molecular markers. The narG and napA gene copy numbers in DNA extracted from 18 different environments showed high variations, with most numbers ranging from 2 x 10(2) to 6.8 x 10(4) copies per ng of DNA. This study provides evidence that, in soil samples, the number of proteobacteria carrying the napA gene is often as high as that of proteobacteria carrying the narG gene. The high correlation observed between narG and napA gene copy numbers in soils suggests that the ecological roles of the corresponding enzymes might be linked.
Figures

References
-
- Berks, B. C., S. J. Ferguson, J. W. B. Moir, and D. J. D. Richardson. 1995. Enzymes and associated electron transports systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1232:97-173. - PubMed
-
- Brunel, B., J. D. Janse, H. J. Laanbroek, and J. D. Woldendorp. 1992. Effect of transient oxic conditions on the composition of the nitrate-reducing community from the rhizosphere of Typha angustipholia. Microb. Ecol. 24:51-61. - PubMed
-
- Chèneby, D., S. Hallet, M. Mondon, F. Martin-Laurent, J. C. Germon, and L. Philippot. 2003. Genetic characterization of the nitrate reducing community based on narG nucleotide sequence analysis. Microb. Ecol. 46:113-121. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

