Concurrent splicing and transcription are not sufficient to enhance splicing efficiency
- PMID: 17630325
- PMCID: PMC1950766
- DOI: 10.1261/rna.595907
Concurrent splicing and transcription are not sufficient to enhance splicing efficiency
Abstract
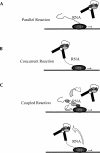
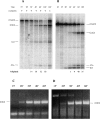
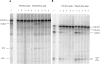
The concept of a tight integration of transcription and splicing of mRNA precursors has been supported with increasing evidence in recent years. However, the mechanism and functional consequences of this integration remain largely unknown. We have examined how these processes impact upon one another when they occur together in HeLa nuclear extract. While both processes do in fact occur in parallel reactions in the extracts, we found no evidence that one process affects the other, under a variety of conditions tested. For example, neither the kinetics nor efficiency of splicing is significantly enhanced by de novo RNA polymerase II-mediated transcription, relative to that of presynthesized RNA added exogenously to the extract. Our results indicate that the act of transcription by RNA polymerase II in vitro is not sufficient to enhance splicing of the newly made RNA.
Figures

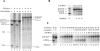
References
-
- Adamson, T.E., Shutt, D.C., Price, D.H. Functional coupling of cleavage and polyadenylation with transcription of mRNA. J. Biol. Chem. 2005;280:32262–32271. - PubMed
-
- Ares, M., Proudfoot, N.J. The Spanish connection: Transcription and mRNA processing get even closer. Cell. 2005;120:163–166. - PubMed
-
- Batsché, E., Yaniv, M., Muchardt, C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2005;13:22–29. - PubMed
-
- Bentley, D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
