Protein-protein interaction hotspots carved into sequences
- PMID: 17630824
- PMCID: PMC1914369
- DOI: 10.1371/journal.pcbi.0030119
Protein-protein interaction hotspots carved into sequences
Abstract
Protein-protein interactions, a key to almost any biological process, are mediated by molecular mechanisms that are not entirely clear. The study of these mechanisms often focuses on all residues at protein-protein interfaces. However, only a small subset of all interface residues is actually essential for recognition or binding. Commonly referred to as "hotspots," these essential residues are defined as residues that impede protein-protein interactions if mutated. While no in silico tool identifies hotspots in unbound chains, numerous prediction methods were designed to identify all the residues in a protein that are likely to be a part of protein-protein interfaces. These methods typically identify successfully only a small fraction of all interface residues. Here, we analyzed the hypothesis that the two subsets correspond (i.e., that in silico methods may predict few residues because they preferentially predict hotspots). We demonstrate that this is indeed the case and that we can therefore predict directly from the sequence of a single protein which residues are interaction hotspots (without knowledge of the interaction partner). Our results suggested that most protein complexes are stabilized by similar basic principles. The ability to accurately and efficiently identify hotspots from sequence enables the annotation and analysis of protein-protein interaction hotspots in entire organisms and thus may benefit function prediction and drug development. The server for prediction is available at http://www.rostlab.org/services/isis.
Conflict of interest statement
Figures

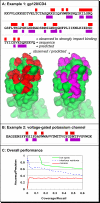
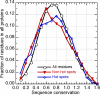

Similar articles
-
PRICE (PRotein Interface Conservation and Energetics): a server for the analysis of protein-protein interfaces.J Struct Funct Genomics. 2011 Mar;12(1):33-41. doi: 10.1007/s10969-011-9108-0. Epub 2011 Apr 26. J Struct Funct Genomics. 2011. PMID: 21519818
-
ISIS: interaction sites identified from sequence.Bioinformatics. 2007 Jan 15;23(2):e13-6. doi: 10.1093/bioinformatics/btl303. Bioinformatics. 2007. PMID: 17237081
-
Exploring the charge space of protein-protein association: a proteomic study.Proteins. 2005 Aug 15;60(3):341-52. doi: 10.1002/prot.20489. Proteins. 2005. PMID: 15887221
-
Characterization and prediction of protein interfaces to infer protein-protein interaction networks.Curr Pharm Biotechnol. 2008 Apr;9(2):67-76. doi: 10.2174/138920108783955191. Curr Pharm Biotechnol. 2008. PMID: 18393863 Review.
-
Interaction-site prediction for protein complexes: a critical assessment.Bioinformatics. 2007 Sep 1;23(17):2203-9. doi: 10.1093/bioinformatics/btm323. Epub 2007 Jun 22. Bioinformatics. 2007. PMID: 17586545 Review.
Cited by
-
Biomacromolecular quantitative structure-activity relationship (BioQSAR): a proof-of-concept study on the modeling, prediction and interpretation of protein-protein binding affinity.J Comput Aided Mol Des. 2013 Jan;27(1):67-78. doi: 10.1007/s10822-012-9625-3. Epub 2013 Jan 10. J Comput Aided Mol Des. 2013. PMID: 23306464
-
Alternative protein-protein interfaces are frequent exceptions.PLoS Comput Biol. 2012;8(8):e1002623. doi: 10.1371/journal.pcbi.1002623. Epub 2012 Aug 2. PLoS Comput Biol. 2012. PMID: 22876170 Free PMC article.
-
On the binding affinity of macromolecular interactions: daring to ask why proteins interact.J R Soc Interface. 2012 Dec 12;10(79):20120835. doi: 10.1098/rsif.2012.0835. Print 2013 Feb. J R Soc Interface. 2012. PMID: 23235262 Free PMC article. Review.
-
Disease-related mutations predicted to impact protein function.BMC Genomics. 2012 Jun 18;13 Suppl 4(Suppl 4):S11. doi: 10.1186/1471-2164-13-S4-S11. BMC Genomics. 2012. PMID: 22759649 Free PMC article.
-
Enabling technology and core theory of synthetic biology.Sci China Life Sci. 2023 Aug;66(8):1742-1785. doi: 10.1007/s11427-022-2214-2. Epub 2023 Feb 6. Sci China Life Sci. 2023. PMID: 36753021 Free PMC article. Review.
References
-
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae . Nature. 2000;403:623–627. - PubMed
-
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. - PubMed
-
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. - PubMed
-
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, et al. A protein interaction map of Drosophila melanogaster . Science. 2003;302:1727–1736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

