The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation
- PMID: 17631636
- PMCID: PMC2045186
- DOI: 10.1128/JB.00023-07
The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation
Abstract
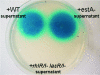
Pseudomonas aeruginosa PAO1 produces the biodetergent rhamnolipid and secretes it into the extracellular environment. The role of rhamnolipids in the life cycle and pathogenicity of P. aeruginosa has not been completely understood, but they are known to affect outer membrane composition, cell motility, and biofilm formation. This report is focused on the influence of the outer membrane-bound esterase EstA of P. aeruginosa PAO1 on rhamnolipid production. EstA is an autotransporter protein which exposes its catalytically active esterase domain on the cell surface. Here we report that the overexpression of EstA in the wild-type background of P. aeruginosa PAO1 results in an increased production of rhamnolipids whereas an estA deletion mutant produced only marginal amounts of rhamnolipids. Also the known rhamnolipid-dependent cellular motility and biofilm formation were affected. Although only a dependence of swarming motility on rhamnolipids has been known so far, the other kinds of motility displayed by P. aeruginosa PAO1, swimming and twitching, were also affected by an estA mutation. In order to demonstrate that EstA enzyme activity is responsible for these effects, inactive variant EstA* was constructed by replacement of the active serine by alanine. None of the mutant phenotypes could be complemented by expression of EstA*, demonstrating that the phenotypes affected by the estA mutation depend on the enzymatically active protein.
Figures
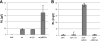



Similar articles
-
PqsR-dependent and PqsR-independent regulation of motility and biofilm formation by PQS in Pseudomonas aeruginosa PAO1.J Basic Microbiol. 2014 Jul;54(7):633-43. doi: 10.1002/jobm.201300091. Epub 2013 Aug 29. J Basic Microbiol. 2014. PMID: 23996096
-
Lipase LipC affects motility, biofilm formation and rhamnolipid production in Pseudomonas aeruginosa.FEMS Microbiol Lett. 2010 Aug 1;309(1):25-34. doi: 10.1111/j.1574-6968.2010.02017.x. Epub 2010 May 17. FEMS Microbiol Lett. 2010. PMID: 20546309
-
Elastase LasB of Pseudomonas aeruginosa promotes biofilm formation partly through rhamnolipid-mediated regulation.Can J Microbiol. 2014 Apr;60(4):227-35. doi: 10.1139/cjm-2013-0667. Epub 2014 Mar 7. Can J Microbiol. 2014. PMID: 24693981
-
Production of rhamnolipids by Pseudomonas aeruginosa.Appl Microbiol Biotechnol. 2005 Oct;68(6):718-25. doi: 10.1007/s00253-005-0150-3. Epub 2005 Oct 13. Appl Microbiol Biotechnol. 2005. PMID: 16160828 Review.
-
Why do microorganisms produce rhamnolipids?World J Microbiol Biotechnol. 2012 Feb;28(2):401-19. doi: 10.1007/s11274-011-0854-8. World J Microbiol Biotechnol. 2012. PMID: 22347773 Free PMC article. Review.
Cited by
-
A membrane-bound esterase PA2949 from Pseudomonas aeruginosa is expressed and purified from Escherichia coli.FEBS Open Bio. 2016 Apr 19;6(5):484-93. doi: 10.1002/2211-5463.12061. eCollection 2016 May. FEBS Open Bio. 2016. PMID: 27419054 Free PMC article.
-
Fucoidan-Stabilized Gold Nanoparticle-Mediated Biofilm Inhibition, Attenuation of Virulence and Motility Properties in Pseudomonas aeruginosa PAO1.Mar Drugs. 2019 Apr 3;17(4):208. doi: 10.3390/md17040208. Mar Drugs. 2019. PMID: 30987163 Free PMC article.
-
Interference with Pseudomonas aeruginosa Quorum Sensing and Virulence by the Mycobacterial Pseudomonas Quinolone Signal Dioxygenase AqdC in Combination with the N-Acylhomoserine Lactone Lactonase QsdA.Infect Immun. 2019 Sep 19;87(10):e00278-19. doi: 10.1128/IAI.00278-19. Print 2019 Oct. Infect Immun. 2019. PMID: 31308081 Free PMC article.
-
Identification of FDA-approved antivirulence drugs targeting the Pseudomonas aeruginosa quorum sensing effector protein PqsE.Virulence. 2020 Dec;11(1):652-668. doi: 10.1080/21505594.2020.1770508. Virulence. 2020. PMID: 32423284 Free PMC article.
-
Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417.BMC Genomics. 2015 Jul 22;16(1):539. doi: 10.1186/s12864-015-1632-z. BMC Genomics. 2015. PMID: 26198432 Free PMC article.
References
-
- Barker, A. P., A. I. Vasil, A. Filloux, G. Ball, P. J. Wilderman, and M. L. Vasil. 2004. A novel extracellular phospholipase C of Pseudomonas aeruginosa is required for phospholipid chemotaxis. Mol. Microbiol. 53:1089-1098. - PubMed
-
- Beal, R., and W. B. Betts. 2000. Role of rhamnolipid biosurfactants in the uptake and mineralization of hexadecane in Pseudomonas aeruginosa. J. Appl. Microbiol. 89:158-168. - PubMed
-
- Becker, S., S. Theile, N. Heppeler, A. Michalczyk, A. Wentzel, S. Wilhelm, K. E. Jaeger, and H. Kolmar. 2005. A generic system for the Escherichia coli cell-surface display of lipolytic enzymes. FEBS Lett. 579:1177-1182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

