Multiple pathways involved in the biosynthesis of anandamide
- PMID: 17631919
- PMCID: PMC2219543
- DOI: 10.1016/j.neuropharm.2007.05.020
Multiple pathways involved in the biosynthesis of anandamide
Abstract
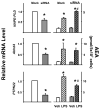
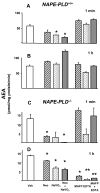
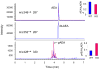
Endocannabinoids, including anandamide (arachidonoyl ethanolamide) have been implicated in the regulation of a growing number of physiological and pathological processes. Anandamide can be generated from its membrane phospholipid precursor N-arachidonoyl phosphatidylethanolamine (NAPE) through hydrolysis by a phospholipase D (NAPE-PLD). Recent evidence indicates, however, the existence of two additional, parallel pathways. One involves the sequential deacylation of NAPE by alpha,beta-hydrolase 4 (Abhd4) and the subsequent cleavage of glycerophosphate to yield anandamide, and the other one proceeds through phospholipase C-mediated hydrolysis of NAPE to yield phosphoanandamide, which is then dephosphorylated by phosphatases, including the tyrosine phosphatase PTPN22 and the inositol 5' phosphatase SHIP1. Conversion of synthetic NAPE to AEA by brain homogenates from wild-type and NAPE-PLD(-/-) mice can proceed through both the PLC/phosphatase and Abdh4 pathways, with the former being dominant at shorter (<10 min) and the latter at longer (60 min) incubations. In macrophages, the endotoxin-induced synthesis of anandamide proceeds uniquely through the phospholipase C/phosphatase pathway.
Figures
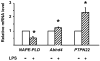

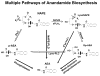
References
-
- An H, Xu H, Zhang M, Zhou J, Feng T, Qian C, Qi R, Cao X. Src homology 2 domain-containing inositol-5-phosphatase 1 (SHIP1) negatively regulates TLR4-mediated LPS reponse primarily through a phosphatase activity- and PI-3K-independent mechanism. Blood. 2005;105:4685–4692. - PubMed
-
- Devane Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949. - PubMed
-
- Di Marzo V, Fontana A, Cadas H, Schinelli S, Cimino G, Schwartz JC, Piomelli D. Formation and inactivation of endogenous cannabinoid anandamide in central neurons. Nature. 1994;372:686–691. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

