HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration
- PMID: 17631931
- PMCID: PMC2140271
- DOI: 10.1016/j.virol.2007.06.001
HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration
Abstract
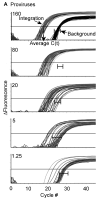
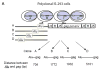
Human Immunodeficiency Virus Type 1 (HIV-1) establishes a latent reservoir early in infection that is resistant to the host immune response and treatment with highly active antiretroviral therapy (HAART). The best understood of these reservoirs forms in resting CD4(+) T cells. While it remains unclear how reservoirs form, a popular model holds that the virus can only integrate in activated CD4(+) T cells. Contrary to this model, our previous results suggest that HIV-1 can integrate directly into the genomes of resting CD4(+) T cells. However, a limitation of our previous studies was that they were conducted at high viral inoculum and these conditions may lead to cellular activation or saturation of restriction factors. In the present study, we tested if our previous findings were an artifact of high inoculum. To do this, we enhanced the sensitivity of our integration assay by incorporating a repetitive sampling technique that allowed us to capture rare integration events that occur near an Alu repeat. The new technique represents a significant advance as it enabled us to measure integration accurately down to 1 provirus/well in 15,000 genomes--a 40-fold enhancement over our prior assay. Using this assay, we demonstrate that HIV can integrate into resting CD4(+) T cells in vitro even at low viral inoculum. These findings suggest there is no threshold number of virions required for HIV to integrate into resting CD4(+) T cells.
Figures



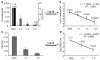


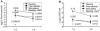
References
-
- Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. - PubMed
-
- Blankson JN, Persaud D, Siliciano RF. The challenge of viral reservoirs in HIV-1 infection. Annual Review of Medicine. 2002;53:557–593. - PubMed
-
- Blumenthal R, Bali-Puri A, Walter A, Covell D, Eidelman O. pH-dependent fusion of vesicular stomatitis virus with Vero cells. Measurement by dequenching of octadecyl rhodamine fluorescence. J Biol Chem. 1987;262(28):13614–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

