How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model
- PMID: 17632573
- PMCID: PMC2766523
- DOI: 10.1038/nrmicro1705
How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model
Abstract
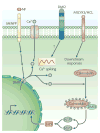
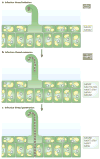
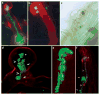
Nitrogen-fixing rhizobial bacteria and leguminous plants have evolved complex signal exchange mechanisms that allow a specific bacterial species to induce its host plant to form invasion structures through which the bacteria can enter the plant root. Once the bacteria have been endocytosed within a host-membrane-bound compartment by root cells, the bacteria differentiate into a new form that can convert atmospheric nitrogen into ammonia. Bacterial differentiation and nitrogen fixation are dependent on the microaerobic environment and other support factors provided by the plant. In return, the plant receives nitrogen from the bacteria, which allows it to grow in the absence of an external nitrogen source. Here, we review recent discoveries about the mutual recognition process that allows the model rhizobial symbiont Sinorhizobium meliloti to invade and differentiate inside its host plant alfalfa (Medicago sativa) and the model host plant barrel medic (Medicago truncatula).
Figures


References
-
- Barnett MJ, Fisher RF. Global gene expression in the rhizobial–legume symbiosis. Symbiosis. 2006;42:1–24.
-
- Oldroyd GE, Downie JA. Calcium, kinases and nodulation signalling in legumes. Nature Rev Mol Cell Biol. 2004;5:566–576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

