Whole genome expression profiling reveals a significant role for immune function in human abdominal aortic aneurysms
- PMID: 17634102
- PMCID: PMC1934369
- DOI: 10.1186/1471-2164-8-237
Whole genome expression profiling reveals a significant role for immune function in human abdominal aortic aneurysms
Abstract
Background: Abdominal aortic aneurysms are a common disorder with an incompletely understood etiology. We used Illumina and Affymetrix microarray platforms to generate global gene expression profiles for both aneurysmal (AAA) and non-aneurysmal abdominal aorta, and identified genes that were significantly differentially expressed between cases and controls.
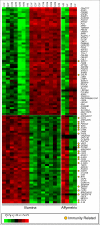
Results: Affymetrix and Illumina arrays included 18,057 genes in common; 11,542 (64%) of these genes were considered to be expressed in either aneurysmal or normal abdominal aorta. There were 3,274 differentially expressed genes with a false discovery rate (FDR) </= 0.05. Many of these genes were not previously known to be involved in AAA, including SOST and RUNX3, which were confirmed using Q-RT-PCR (Pearson correlation coefficient for microarray and Q-RT-PCR data = 0.89; p-values for differences in expression between AAA and controls for SOST: 4.87 x 10-4 and for RUNX3: 4.33 x 10-5). Analysis of biological pathways, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG), indicated extreme overrepresentation of immune related categories. The enriched categories included the GO category Immune Response (GO:0006955; FDR = 2.1 x 10-14), and the KEGG pathways natural killer cell mediated cytotoxicity (hsa04650; FDR = 5.9 x 10-6) and leukocyte transendothelial migration (hsa04670; FDR = 1.1 x 10-5).
Conclusion: Previous studies have provided evidence for the involvement of the immune system in AAA. The current expression analysis extends these findings by demonstrating broad coordinate gene expression in immunological pathways. A large number of genes involved in immune function were differentially expressed in AAA, and the pathway analysis gave these results a biological context. The data provide valuable insight for future studies to dissect the pathogenesis of human AAA. These pathways might also be used as targets for the development of therapeutic agents for AAA.
Figures


Similar articles
-
Transcriptional (ChIP-Chip) Analysis of ELF1, ETS2, RUNX1 and STAT5 in Human Abdominal Aortic Aneurysm.Int J Mol Sci. 2015 May 18;16(5):11229-58. doi: 10.3390/ijms160511229. Int J Mol Sci. 2015. PMID: 25993293 Free PMC article.
-
Microarray-based Gene Expression Profiling of Abdominal Aortic Aneurysm.Eur J Vasc Endovasc Surg. 2016 Jul;52(1):47-55. doi: 10.1016/j.ejvs.2016.03.016. Epub 2016 May 3. Eur J Vasc Endovasc Surg. 2016. PMID: 27157464
-
Global expression profiles in human normal and aneurysmal abdominal aorta based on two distinct whole genome microarray platforms.Ann N Y Acad Sci. 2006 Nov;1085:360-2. doi: 10.1196/annals.1383.041. Ann N Y Acad Sci. 2006. PMID: 17182956
-
Exome sequencing reveals new insights into the progression of abdominal aortic aneurysm.Eur Rev Med Pharmacol Sci. 2013 Sep;17(17):2401-9. Eur Rev Med Pharmacol Sci. 2013. PMID: 24065236 Review.
-
The genetic basis for aortic aneurysmal disease.Heart. 2014 Jun;100(12):916-22. doi: 10.1136/heartjnl-2013-305130. Heart. 2014. PMID: 24842835 Review.
Cited by
-
Transcriptional (ChIP-Chip) Analysis of ELF1, ETS2, RUNX1 and STAT5 in Human Abdominal Aortic Aneurysm.Int J Mol Sci. 2015 May 18;16(5):11229-58. doi: 10.3390/ijms160511229. Int J Mol Sci. 2015. PMID: 25993293 Free PMC article.
-
Multiset sparse partial least squares path modeling for high dimensional omics data analysis.BMC Bioinformatics. 2020 Jan 9;21(1):9. doi: 10.1186/s12859-019-3286-3. BMC Bioinformatics. 2020. PMID: 31918677 Free PMC article.
-
Differentially expressed genes and canonical pathways in the ascending thoracic aortic aneurysm - The Tampere Vascular Study.Sci Rep. 2017 Sep 21;7(1):12127. doi: 10.1038/s41598-017-12421-4. Sci Rep. 2017. PMID: 28935963 Free PMC article.
-
Identification of PTPN22 as a potential genetic biomarker for abdominal aortic aneurysm.Front Cardiovasc Med. 2022 Dec 14;9:1061771. doi: 10.3389/fcvm.2022.1061771. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36588574 Free PMC article.
-
RUNX3 is up-regulated in abdominal aortic aneurysm and regulates the function of vascular smooth muscle cells by regulating TGF-β1.J Mol Histol. 2022 Feb;53(1):1-11. doi: 10.1007/s10735-021-10035-9. Epub 2021 Nov 23. J Mol Histol. 2022. PMID: 34813022
References
-
- Alcorn HG, Wolfson SK, Jr., Sutton-Tyrrell K, Kuller LH, O'Leary D. Risk factors for abdominal aortic aneurysms in older adults enrolled in The Cardiovascular Health Study. Arterioscler Thromb Vasc Biol. 1996;16:963–970. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

