RASSF1A promoter methylation and expression analysis in normal and neoplastic kidney indicates a role in early tumorigenesis
- PMID: 17634119
- PMCID: PMC1939711
- DOI: 10.1186/1476-4598-6-49
RASSF1A promoter methylation and expression analysis in normal and neoplastic kidney indicates a role in early tumorigenesis
Abstract
Background: Epigenetic silencing of the RAS association domain family 1A (RASSF1A) tumor suppressor gene promoter has been demonstrated in renal cell carcinoma (RCC) as a result of promoter hypermethylation. Contradictory results have been reported for RASSF1A methylation in normal kidney, thus it is not clear whether a significant difference between RASSF1A methylation in normal and tumor cells of the kidney exists. Moreover, RASSF1A expression has not been characterized in tumors or normal tissue as yet.
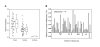
Results: Using combined bisulfite restriction analysis (COBRA) we compared RASSF1A methylation in 90 paired tissue samples obtained from primary kidney tumors and corresponding normal tissue. Bisulfite sequence analysis was carried out using both pooled amplicons from the tumor and normal tissue groups and subclones obtained from a single tissue pair. Expression of RASSF1A was analyzed by the use of tissue arrays and immunohistochemistry. We found significantly increased methylation in tumor samples (mean methylation, 20%) compared to corresponding normal tissues (mean methylation, 11%; P < 0.001). Densely methylated sequences were found both in pooled and individual sequences of normal tissue. Immunohistochemical analysis revealed a significant reduced expression of RASSF1A in most of the tumor samples. Heterogeneous expression patterns of RASSF1A were detected in a subgroup of histologically normal tubular epithelia.
Conclusion: Our methylation and expression data support the hypothesis that RASSF1A is involved in early tumorigenesis of renal cell carcinoma.
Figures





References
-
- Landis SH, Murray T, Bolden S, Wingo PA. Cancer statistics, 1999. CA Cancer J Clin. 1999;49:8–31, 1. - PubMed
-
- Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. - PubMed
-
- Thoenes W, Storkel S, Rumpelt HJ. Histopathology and classification of renal cell tumors (adenomas, oncocytomas and carcinomas). The basic cytological and histopathological elements and their use for diagnostics. Pathol Res Pract. 1986;181:125–143. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

