TR1.3 viral pathogenesis and syncytium formation are linked to Env-Gag cooperation
- PMID: 17634219
- PMCID: PMC2045439
- DOI: 10.1128/JVI.00816-07
TR1.3 viral pathogenesis and syncytium formation are linked to Env-Gag cooperation
Abstract
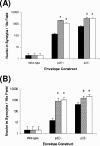
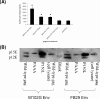
Infection with murine leukemia virus (MLV) TR1.3 or the related molecular construct W102G causes severe neuropathology in vivo. Infection is causally linked to the development of extensive syncytia in brain capillary endothelial cells (BCEC). These viruses also induce cell fusion of murine cell lines, such as SC-1 and NIH 3T3, which are otherwise resistant to MLV-induced syncytium formation. Although the virulence of these viruses maps within the env gene, the mechanism of fusion enhancement is not fully determined. To this end, we examined the capacity of the syncytium-inducing (SI) TR1.3 and W102G MLVs to overcome the fusion inhibitory activity inherent in the full-length Env cytoplasmic tail. These studies showed that the TR1.3 and W102G Envs did not induce premature cleavage of p2E, nor did they override p2E fusion inhibition. Indeed, in the presence of mutations that disrupt p2E function, the TR1.3 and W102G Envs significantly increased the extent of cell fusion compared to that with the non-syncytium-inducing MLV FB29. Surprisingly, we also observed that TR1.3 and W102G Envs failed to elicit syncytium formation in these in vitro assays. Coexpression of gag-pol with env restored syncytium formation, and accordingly, mutations within gag-pol were used to examine the minimal functional requirements for the SI phenotype. The results indicate that both gag-dependent particle budding and cleavage of p2E are required to activate the SI phenotype of TR1.3 and W102G viruses. Collectively, these data suggest that the TR1.3 and W102G viruses induce cell fusion by the fusion-from-without pathway.
Figures




Similar articles
-
Induction of syncytia by neuropathogenic murine leukemia viruses depends on receptor density, host cell determinants, and the intrinsic fusion potential of envelope protein.J Virol. 1999 Nov;73(11):9377-85. doi: 10.1128/JVI.73.11.9377-9385.1999. J Virol. 1999. PMID: 10516046 Free PMC article.
-
Neuropathogenic murine leukemia virus TR1.3 induces selective syncytia formation of brain capillary endothelium.Virology. 2004 Mar 30;321(1):57-64. doi: 10.1016/j.virol.2003.12.002. Virology. 2004. PMID: 15033565
-
Intracellular trafficking of Gag and Env proteins and their interactions modulate pseudotyping of retroviruses.J Virol. 2004 Jul;78(13):7153-64. doi: 10.1128/JVI.78.13.7153-7164.2004. J Virol. 2004. PMID: 15194792 Free PMC article.
-
Synthesis and assembly of chimeric human immunodeficiency virus gag pseudovirions.Intervirology. 1996;39(1-2):40-8. doi: 10.1159/000150473. Intervirology. 1996. PMID: 8957668 Review.
-
Placenta trophoblast fusion.Methods Mol Biol. 2008;475:135-47. doi: 10.1007/978-1-59745-250-2_8. Methods Mol Biol. 2008. PMID: 18979242 Review.
Cited by
-
Quantitative assays reveal cell fusion at minimal levels of SARS-CoV-2 spike protein and fusion from without.iScience. 2021 Mar 19;24(3):102170. doi: 10.1016/j.isci.2021.102170. Epub 2021 Feb 9. iScience. 2021. PMID: 33585805 Free PMC article.
-
A mutant retroviral receptor restricts virus superinfection interference and productive infection.Retrovirology. 2012 Jun 12;9:51. doi: 10.1186/1742-4690-9-51. Retrovirology. 2012. PMID: 22691439 Free PMC article.
-
Unique N-linked glycosylation of CasBrE Env influences its stability, processing, and viral infectivity but not its neurotoxicity.J Virol. 2013 Aug;87(15):8372-87. doi: 10.1128/JVI.00392-13. Epub 2013 May 22. J Virol. 2013. PMID: 23698308 Free PMC article.
References
-
- Alimonti, J. B., T. B. Ball, and K. R. Fowke. 2003. Mechanisms of CD4+ T lymphocyte cell death in human immunodeficiency virus infection and AIDS. J. Gen. Virol. 84:1649-1661. - PubMed
-
- Alin, K., and S. P. Goff. 1996. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology 222:339-351. - PubMed
-
- Alin, K., and S. P. Goff. 1996. Mutational analysis of interactions between the Gag precursor proteins of murine leukemia viruses. Virology 216:418-424. - PubMed
-
- Andreau, K., J. L. Perfettini, M. Castedo, D. Metivier, V. Scott, G. Pierron, and G. Kroemer. 2004. Contagious apoptosis facilitated by the HIV-1 envelope: fusion-induced cell-to-cell transmission of a lethal signal. J. Cell Sci. 117:5643-5653. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

