Herpes simplex virus type 1 C-terminal variants of the origin binding protein (OBP), OBPC-1 and OBPC-2, cooperatively regulate viral DNA levels in vitro, and OBPC-2 affects mortality in mice
- PMID: 17634223
- PMCID: PMC2045454
- DOI: 10.1128/JVI.01213-07
Herpes simplex virus type 1 C-terminal variants of the origin binding protein (OBP), OBPC-1 and OBPC-2, cooperatively regulate viral DNA levels in vitro, and OBPC-2 affects mortality in mice
Abstract
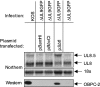
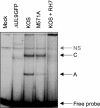
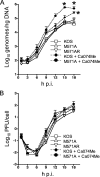
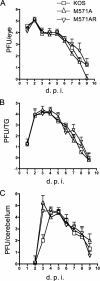
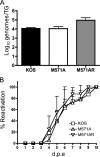
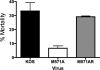
Two in-frame, C-terminal isoforms of the herpes simplex virus type 1 (HSV-1) origin binding protein (OBP), OBPC-1 and OBPC-2, and a unique C-terminal transcript, UL8.5, are specified by HSV-1 DNA. As the first isoform identified, OBPC-1 was initially assumed to be the product of the UL8.5 transcript. Recent evidence has demonstrated, however, that OBPC-1 is a cathepsin B-mediated cleavage product of OBP, suggesting that OBPC-2 is the product of the UL8.5 transcript. Because both OBPC-1 and -2 contain the majority of the OBP DNA binding domain, we hypothesized that both may be involved in regulating origin-dependent, OBP-mediated viral DNA replication. In this paper, we demonstrate that OBPC-2 is, indeed, the product of the UL8.5 transcript. The translational start site of OBPC-2 was mapped, and a virus (M571A) that does not express this protein efficiently was constructed. Using M571A, we have shown that OBPC-2 is able to bind origin DNA, even though it lacks seven N-terminal amino acid residues of the previously mapped OBP DNA binding domain, resulting in a revision of the limits of the OBP DNA binding domain. Consistent with their proposed roles in regulating viral DNA replication, OBPC-1 and -2 act together to down-regulate viral DNA replication in vitro. During functional studies in vivo, OBPC-2 was identified as a factor that increases mortality in the mouse ocular model of HSV-1 infection.
Figures



Similar articles
-
Cathepsin B mediates cleavage of herpes simplex virus type 1 origin binding protein (OBP) to yield OBPC-1, and cleavage is dependent upon viral DNA replication.J Virol. 2007 Sep;81(17):9175-82. doi: 10.1128/JVI.00676-07. Epub 2007 Jun 6. J Virol. 2007. PMID: 17553869 Free PMC article.
-
Origin binding protein-containing protein-DNA complex formation at herpes simplex virus type 1 oriS: role in oriS-dependent DNA replication.J Virol. 2001 Aug;75(15):6808-16. doi: 10.1128/JVI.75.15.6808-6816.2001. J Virol. 2001. PMID: 11435559 Free PMC article.
-
Transcriptional analysis of the region of the herpes simplex virus type 1 genome containing the UL8, UL9, and UL10 genes and identification of a novel delayed-early gene product, OBPC.J Virol. 1994 Jul;68(7):4251-61. doi: 10.1128/JVI.68.7.4251-4261.1994. J Virol. 1994. PMID: 8207800 Free PMC article.
-
Herpes simplex virus type 1 replication and recombination.Biochimie. 1995;77(10):787-95. doi: 10.1016/0300-9084(96)88197-1. Biochimie. 1995. PMID: 8824776 Review.
-
Replication of herpes simplex virus DNA.J Biol Chem. 1999 Oct 1;274(40):28059-62. doi: 10.1074/jbc.274.40.28059. J Biol Chem. 1999. PMID: 10497152 Review. No abstract available.
Cited by
-
Direct interaction between the N- and C-terminal portions of the herpes simplex virus type 1 origin binding protein UL9 implies the formation of a head-to-tail dimer.J Virol. 2007 Dec;81(24):13659-67. doi: 10.1128/JVI.01204-07. Epub 2007 Oct 17. J Virol. 2007. PMID: 17942532 Free PMC article.
-
Replication and recombination of herpes simplex virus DNA.J Biol Chem. 2011 May 6;286(18):15619-24. doi: 10.1074/jbc.R111.233981. Epub 2011 Mar 1. J Biol Chem. 2011. PMID: 21362621 Free PMC article. Review.
-
A sequence within the varicella-zoster virus (VZV) OriS is a negative regulator of DNA replication and is bound by a protein complex containing the VZV ORF29 protein.J Virol. 2011 Dec;85(23):12188-200. doi: 10.1128/JVI.05501-11. Epub 2011 Sep 21. J Virol. 2011. PMID: 21937644 Free PMC article.
-
Herpes simplex virus 1 ICP0 phosphorylation site mutants are attenuated for viral replication and impaired for explant-induced reactivation.J Virol. 2011 Dec;85(23):12631-7. doi: 10.1128/JVI.05661-11. Epub 2011 Sep 21. J Virol. 2011. PMID: 21937654 Free PMC article.
-
Role of a cdk5-associated protein, p35, in herpes simplex virus type 1 replication in vivo.J Neurovirol. 2010 Oct;16(5):405-9. doi: 10.3109/13550284.2010.513030. J Neurovirol. 2010. PMID: 20839922 Free PMC article.
References
-
- Arbuckle, M. I., and N. D. Stow. 1993. A mutational analysis of the DNA-binding domain of the herpes simplex virus type 1 UL9 protein. J. Gen. Virol. 74:1349-1355. - PubMed
-
- Balliet, J. W., J. C. Min, M. S. Cabatingan, and P. A. Schaffer. 2005. Site-directed mutagenesis of large DNA palindromes: construction and in vitro characterization of herpes simplex virus type 1 mutants containing point mutations that eliminate the oriL or oriS initiation function. J. Virol. 79:12783-12797. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

