A tyrosine-based signal plays a critical role in the targeting and function of adenovirus RIDalpha protein
- PMID: 17634224
- PMCID: PMC2045482
- DOI: 10.1128/JVI.00399-07
A tyrosine-based signal plays a critical role in the targeting and function of adenovirus RIDalpha protein
Abstract
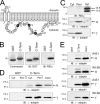
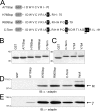
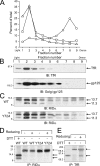
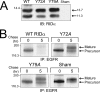
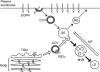
Early region 3 genes of human adenoviruses contribute to the virus life cycle by altering the trafficking of cellular proteins involved in adaptive immunity and inflammatory responses. The ability of early region 3 genes to target specific molecules suggests that they could be used to curtail pathological processes associated with these molecules and treat human disease. However, this approach requires genetic dissection of the multiple functions attributed to early region 3 genes. The purpose of this study was to determine the role of targeting on the ability of the early region 3-encoded protein RIDalpha to downregulate the EGF receptor. A fusion protein between the RIDalpha cytoplasmic tail and glutathione S-transferase was used to isolate clathrin-associated adaptor 1 and adaptor 2 protein complexes from mammalian cells. Deletion and site-directed mutagenesis studies showed that residues 71-AYLRH of RIDalpha are necessary for in vitro binding to both adaptor complexes and that Tyr72 has an important role in these interactions. In addition, RIDalpha containing a Y72A point mutation accumulates in the trans-Golgi network and fails to downregulate the EGF receptor when it is introduced into mammalian cells as a transgene. Altogether, our data suggest a model where RIDalpha is trafficked directly from the trans-Golgi network to an endosomal compartment, where it intercepts EGF receptors undergoing constitutive recycling to the plasma membrane and redirects them to lysosomes.
Figures



Similar articles
-
Adenovirus early region 3 RIDα protein limits NFκB signaling through stress-activated EGF receptors.PLoS Pathog. 2019 Aug 19;15(8):e1008017. doi: 10.1371/journal.ppat.1008017. eCollection 2019 Aug. PLoS Pathog. 2019. PMID: 31425554 Free PMC article.
-
Adenovirus RIDalpha regulates endosome maturation by mimicking GTP-Rab7.J Cell Biol. 2007 Dec 3;179(5):965-80. doi: 10.1083/jcb.200702187. Epub 2007 Nov 26. J Cell Biol. 2007. PMID: 18039930 Free PMC article.
-
E3-13.7 integral membrane proteins encoded by human adenoviruses alter epidermal growth factor receptor trafficking by interacting directly with receptors in early endosomes.Mol Biol Cell. 2000 Oct;11(10):3559-72. doi: 10.1091/mbc.11.10.3559. Mol Biol Cell. 2000. PMID: 11029055 Free PMC article.
-
Traffic Through the Trans-Golgi Network and the Endosomal System Requires Collaboration Between Exomer and Clathrin Adaptors in Fission Yeast.Genetics. 2017 Feb;205(2):673-690. doi: 10.1534/genetics.116.193458. Epub 2016 Dec 14. Genetics. 2017. PMID: 27974503 Free PMC article.
-
New Insights to Adenovirus-Directed Innate Immunity in Respiratory Epithelial Cells.Microorganisms. 2019 Jul 25;7(8):216. doi: 10.3390/microorganisms7080216. Microorganisms. 2019. PMID: 31349602 Free PMC article. Review.
Cited by
-
Autosomal recessive polycystic kidney disease epithelial cell model reveals multiple basolateral epidermal growth factor receptor sorting pathways.Mol Biol Cell. 2010 Aug 1;21(15):2732-45. doi: 10.1091/mbc.e09-12-1059. Epub 2010 Jun 2. Mol Biol Cell. 2010. PMID: 20519437 Free PMC article.
-
Human Adenoviruses, Cholesterol Trafficking, and NF-κB Signaling.J Immunol Sci. 2018;2(1):9-14. Epub 2018 Jan 16. J Immunol Sci. 2018. PMID: 30090876 Free PMC article.
-
Adenovirus Modulates Toll-Like Receptor 4 Signaling by Reprogramming ORP1L-VAP Protein Contacts for Cholesterol Transport from Endosomes to the Endoplasmic Reticulum.J Virol. 2017 Feb 28;91(6):e01904-16. doi: 10.1128/JVI.01904-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28077646 Free PMC article.
-
Adenovirus RID-alpha activates an autonomous cholesterol regulatory mechanism that rescues defects linked to Niemann-Pick disease type C.J Cell Biol. 2009 Nov 16;187(4):537-52. doi: 10.1083/jcb.200903039. J Cell Biol. 2009. PMID: 19948501 Free PMC article.
-
Adenovirus early region 3 RIDα protein limits NFκB signaling through stress-activated EGF receptors.PLoS Pathog. 2019 Aug 19;15(8):e1008017. doi: 10.1371/journal.ppat.1008017. eCollection 2019 Aug. PLoS Pathog. 2019. PMID: 31425554 Free PMC article.
References
-
- Avila, M. M., G. Carballal, H. Rovaletti, B. Ebekian, M. Cusminsky, and M. Weissenbacher. 1989. Viral etiology in acute lower respiratory infections in children from a closed community. Am. Rev. Respir. Dis. 140:634-637. - PubMed
-
- Bonifacino, J., and L. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395-447. - PubMed
-
- Brandt, C. D., H. W. Kim, A. J. Vargosko, B. C. Jeffries, J. O. Arrobio, B. Rindge, R. H. Parrott, and R. M. Chanock. 1969. Infections in 18,000 infants and children in a controlled study of respiratory tract disease. I. Adenovirus pathogenicity in relation to serologic type and illness syndrome. Am. J. Epidemiol. 90:484-500. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

