Modification of small RNAs associated with suppression of RNA silencing by tobamovirus replicase protein
- PMID: 17634237
- PMCID: PMC2045474
- DOI: 10.1128/JVI.00727-07
Modification of small RNAs associated with suppression of RNA silencing by tobamovirus replicase protein
Abstract
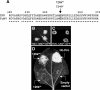
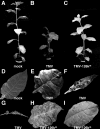
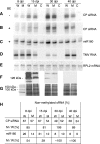
Plant viruses act as triggers and targets of RNA silencing and have evolved proteins to suppress this plant defense response during infection. Although Tobacco mosaic tobamovirus (TMV) triggers the production of virus-specific small interfering RNAs (siRNAs), this does not lead to efficient silencing of TMV nor is a TMV-green fluorescent protein (GFP) hybrid able to induce silencing of a GFP-transgene in Nicotiana benthamiana, indicating that a TMV silencing suppressor is active and acts downstream of siRNA production. On the other hand, TMV-GFP is unable to spread into cells in which GFP silencing is established, suggesting that the viral silencing suppressor cannot revert silencing that is already established. Although previous evidence indicates that the tobamovirus silencing suppressing activity resides in the viral 126-kDa small replicase subunit, the mechanism of silencing suppression by this virus family is not known. Here, we connect the silencing suppressing activity of this protein with our previous finding that Oilseed rape mosaic tobamovirus infection leads to interference with HEN1-mediated methylation of siRNA and micro-RNA (miRNA). We demonstrate that TMV infection similarly leads to interference with HEN1-mediated methylation of small RNAs and that this interference and the formation of virus-induced disease symptoms are linked to the silencing suppressor activity of the 126-kDa protein. Moreover, we show that also Turnip crinkle virus interferes with the methylation of siRNA but, in contrast to tobamoviruses, not with the methylation of miRNA.
Figures




Similar articles
-
Revisiting the Roles of Tobamovirus Replicase Complex Proteins in Viral Replication and Silencing Suppression.Mol Plant Microbe Interact. 2018 Jan;31(1):125-144. doi: 10.1094/MPMI-07-17-0164-R. Epub 2017 Nov 15. Mol Plant Microbe Interact. 2018. PMID: 29140168
-
The benyvirus RNA silencing suppressor is essential for long-distance movement, requires both zinc-finger and NoLS basic residues but not a nucleolar localization for its silencing-suppression activity.Mol Plant Microbe Interact. 2013 Feb;26(2):168-81. doi: 10.1094/MPMI-06-12-0142-R. Mol Plant Microbe Interact. 2013. PMID: 23013437
-
The Tobacco mosaic virus 126-kDa protein associated with virus replication and movement suppresses RNA silencing.Mol Plant Microbe Interact. 2004 Jun;17(6):583-92. doi: 10.1094/MPMI.2004.17.6.583. Mol Plant Microbe Interact. 2004. PMID: 15195941
-
Modulation of host plant immunity by Tobamovirus proteins.Ann Bot. 2017 Mar 1;119(5):737-747. doi: 10.1093/aob/mcw216. Ann Bot. 2017. PMID: 27941090 Free PMC article. Review.
-
Control of tobamovirus infections via pathogen-derived resistance.Adv Virus Res. 1999;53:369-86. doi: 10.1016/s0065-3527(08)60357-7. Adv Virus Res. 1999. PMID: 10582108 Review. No abstract available.
Cited by
-
The 35-amino acid C2 protein of Cotton leaf curl Kokhran virus, Burewala, implicated in resistance breaking in cotton, retains some activities of the full-length protein.Virus Genes. 2016 Oct;52(5):688-97. doi: 10.1007/s11262-016-1357-3. Epub 2016 May 21. Virus Genes. 2016. PMID: 27209537
-
Molecular characterization and In Vitro synthesis of infectious RNA of a Turnip vein-clearing virus isolated from Alliaria petiolata in Hungary.PLoS One. 2019 Oct 24;14(10):e0224398. doi: 10.1371/journal.pone.0224398. eCollection 2019. PLoS One. 2019. PMID: 31648277 Free PMC article.
-
RNA-based antiviral immunity.Nat Rev Immunol. 2010 Sep;10(9):632-44. doi: 10.1038/nri2824. Epub 2010 Aug 13. Nat Rev Immunol. 2010. PMID: 20706278 Review.
-
Expression of artificial microRNAs in tomato confers efficient and stable virus resistance in a cell-autonomous manner.Transgenic Res. 2011 Jun;20(3):569-81. doi: 10.1007/s11248-010-9440-3. Epub 2010 Sep 14. Transgenic Res. 2011. PMID: 20835923
-
Plant miRNAome and antiviral resistance: a retrospective view and prospective challenges.Virus Genes. 2014 Feb;48(1):1-14. doi: 10.1007/s11262-014-1038-z. Epub 2014 Jan 21. Virus Genes. 2014. PMID: 24445902 Review.
References
-
- Anandalakshmi, R., R. Marathe, X. Ge, J. M. Herr, Jr., C. Mau, A. Mallory, G. Pruss, L. Bowman, and V. B. Vance. 2000. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290:142-144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

